▻ Overview
▻ Stage Description
▻ Images
▻ Download Supplemental Table
▻ In Situ Hybridization Data
▻ Sequences
Overview
A stage of Smed embryonic development defined by a unique gene expression signature and morphology, 2 - 3 days post-egg capsule deposition at 20C. Sphere formation. A fraction of the blastomeres differentiate into temporary embryonic tissues that provide form and function to the embryo. Undifferentiated blastomeres remain in the embryonic wall. [PMID:28072387]
Stage Description
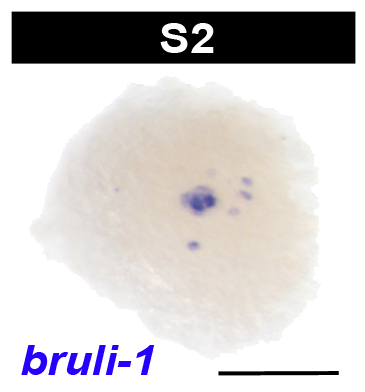
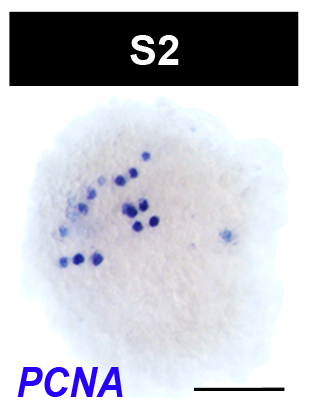
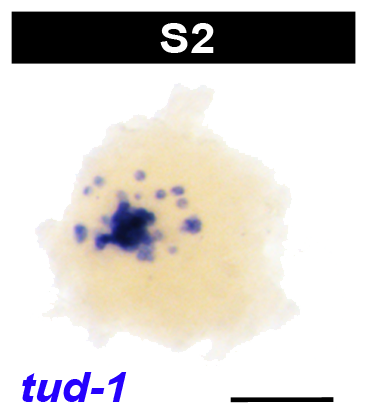
Embryos undergoing the dynamic process of sphere formation are called protospheres, and appear as loosely adherent masses of shaggy yolk, roughly 200 µm in diameter, in live preparations. During sphere formation some blastomeres differentiate into temporary embryonic tissues, providing form and function to the embryo. The primitive ectoderm forms a single cell layer bounding the sphere. Other blastomeres differentiate into the temporary embryonic pharynx and associated primitive gut cells. The temporary embryonic pharynx is an innervated, muscular pump that ingests yolk into the primitive gut cavity. The primitive gut consists of phagocytic cells associated with the temporary embryonic pharynx. Some undifferentiated blastomeres remain in the embryonic wall, the parenchymal space between the primitive gut and ectoderm.
Images


Download Supplemental Table
Figure 1 – source data 2. S2 enriched transcripts from pairwise and/or mixed stage reference comparisons
Criteria for flagging differentially expressed transcripts:
1) Pairwise comparison (S2 vs Y): adjusted p-value < 1e-5 S2 vs mixed reference comparison: adjusted p-value <1e-5 2) log2 ratio ≥ 2.322 (5-fold upregulation, both comparisons) or log2 ratio ≤ -2.322 (5 fold downregulation, pairwise comparison only) 3) Average scaled RPKM value ≥ 1.0 for S2 4) Transcript has ≥ 1 ORF 5) Transcripts derived from transposase or retroviruses were omitted, as were S2 enriched transcripts also upregulated in yolk in the mixed reference comparison.
Tabs in this excel file contain 1) pairwise comparison data (if applicable), 2) mixed stage reference comparison data, 3) cluster membership, average RPKM values across embryogenesis (Y-S8), and in C4 and SX adults, as well as best BLASTx hits (E < 0.001) versus the NR, Swiss-Prot, C. elegans, D. melanogaster, D. rerio, X. tropicalis, M. musculus and H. sapiens REF-Seq databases, 4) GO analysis: Manually curated and categorized Biological Process (BP) GO IDs and 5) GO analysis: unabridged results. S2-S8: Stages 2-8. Y: Yolk. C4: C4 asexual adults. SX: Mature sexual adults
In Situ Hybridization Data
| Abbreviation or symbol | Definition |
|---|---|
| O | oral hemisphere |
| A | aboral hemisphere |
| D | dorsal |
| V | ventral |
| L | lateral |
| black arrowhead | embryonic pharynx |
| red arrowhead | definitive pharynx |
| black arrows | primitive gut |
| yellow arrows | primitive ectoderm cells |
| cyan arrows | brain |
| cyan arrowheads | nerve cords |
| blue arrowheads | eye progenitors (trail cells) |
| purple arrowheads | eyes |
| scale bar | 100 µm |
>SMED30007406 ID=SMED30007406|Name=SMEDWI-1|organism=Schmidtea mediterranea sexual|type=transcript|length=2730bp
AATCGAATACATAAAAATTTATTTGACACCAAATACAAAGAGACATTAATCTACAAGTAAAATAAGCGTTCGCGAATTCT
GTCATTTATCGCTGGAGGAGTAACACCACGATGAATCTTGCCAACGAGTTCAGCCAAACGATGGGCATAATGCGTTGGAA
CAGGAACTCGGATTGTTCCCATCCAGTTAAAATACAAATGTGTTAACGAATAAGTTATTTGCTGCAATACACTCGGAGAC
ATAACTGAAACCTCATTTGTTTTCTTGTTTGTAAACTTAGTGTCCATAAGAACATTATAATTCGTAGGAGATGCTGTACC
TTTAGTTGTTTTCTGAGACACCAAGTAAAATTCATAGAAATTGGGTTTCACTATTTTCTCATCAACCACAGTACCAGGAT
TTGGATTAGCCCCATCTTTGAAAAATTTAACACTAATTCTCTTCTTAACCACAACATAAATAATTTGGGGCAAAGTTTGA
TTTTCGTAAATTTTCTCTATCATTTTCATAACAGCATCAGTTTCGAACTTTTTGGTAAATGCTAACTGTGAATCTCCAAC
ACCGTCACGATAAATAATCAAACGAAGAGGCATAGTGTTAAATTTGTTTTGGAATGTTGTAAGGGCCGTTAGGAAGTTTT
TGCCCAAATTTTCGTGAAATTCATTTTTACCTTTAGATGAATTGACAAAACTGATATACTGAGAAAATTTGGCACTAATT
GAAAATACGGATGCTTGAACAGATCTTTTACCGGTTTTGCTATGAAAAGTATCCAATCCTACAATCATAGTCGGTGCCAT
TTTAAGATTAATACCCCATGGGTCGTATCCTAATTTAGAAAACATTTGCATAACTGATTTATCTGCAACCGTTTTACGAC
GTCGATCATCTCTATTACTTCCGTTTCTCTGCGTCACACACTGAGTCAATAGACCTGTAGACATTGTAAAATTCTTTACT
TTTGCGTAAACTTTATCATCAGGAATGAACACTAATGCCATATGTACCTTGCTTACACGAATAAAATCTGTTAAAGCGTC
TTCAACTTCATTGGATCGACATGAAGATATACTGTCCATTGTGTAATTGATTCTTAATCTTCCTATTTCTCTTTTAACAT
CTTCAATAAAATTGTTGAAGTGACTAGGGTTTCGATCAACAACCAGTACACCAAAACGATGAGCTGTGCCTTTAGGAACG
TCAAATTTAATTTCCCCAAACTTCCAATCATCAGGGATTTTAGTATTAATTTTAACTTCGTTTGAAAATATATCAACTGG
AGCCAATTCACGACCACGAATCGTAATCGGTTGTTCTTCAATACACATCCCCCAGCTCTTCATATAATCTTTTGATTTCC
CAGCTCCGGTCCACTTACAAAAATCTCTTATGTCATTTAATCGTTCTCTAGGCTCACGTTTCAATACGGTTCCTAAATTC
TTTTGCAAATTTATATTACTTCTTTCACTATCCGAAAATCCACATAAATAGCATAATTCACCTGGAATTGATAAACTTTG
ATCTGTTTCTGTAGGTCCTTCAGGTTCATTTTCAGTTTTTTTCTTTTGGGCGAACCATCGATCGCTTGACTCGCGATATA
ACAAATGGTTGTTTGTCATTTTTCAAATTAATGTTATACCGTTGCTTGAAATAATTCAAATAGGATATCTTTTTATCGCC
TAGCTGAACCTCGTCATTAACATCCATTTCTTTAATATCTGAAATTCTATACGTCTTGTTATTATACTTGGTAATTATGT
CCCTTCCTATTAATTGATCAATTTGATCTCGTCGATACACTGATACTACATTGTTTTCATTTAGAACACGATTAATTCTT
TCAAGATAAAGTAAAGTCGAAGCAGTTCCATTTTCATTTGTTCCTCTTGTAATTGTAGTAGAATACCCTCCTAATATCTT
GAAATTGTCCATTTTAAAAAAGCTAGGTCCTTCTCGAATACCACCACTTTCACAATTTGATCCCAGAAAAAATTGTCTAC
CAATTTTTTCTTGTCCCATAAATATTTGAATATTATTAAATACAATATTTATCATCTGATAATATTCTTCAGAATTTTTT
AAAAACGTCGTCAGCAATCGTATGGAAATTTCAACATCTTTGATAACTAAGTTTTCTGTATCTTCATCTGAGTGCCACTT
AGAAGTCGTAAATAATCTTCTGCCATCAAAACAAAATGGTTGACCATTATTTTTGCTGTTTGCATAAGCTAATAATAACT
CTAGCTTAGTACGGATACTTTTATTCTCCATAACTACATCATACATGTAAAATGGTCCTTCGACTTTAATATTAAAGTTA
GCGTAGTTAGACTTGAGACTAACTACTCTTCCATCTTTCCCTATTTTGTCTGTAACTGTATCAGGCTGTAAAGCAGGTTC
AATAATTGCGAATCTTCTTTGGAGACCTCCTCTTCTTGGTCTTAAATTGGGATCACTTGTAACATTAGTTGAATCCATTT
CGTAAATTTGAGTAAAAATGTTAAAACAAAGACTTCGGTTTAAATAAACAAAATATACTATGTAAAACGATAAATATAAT
TGCTTTAATGGTTTATGATTTGTATTATATAATAATAATAATTTAATTTCCCTGTGAATATTATATTAAAATATAGTAAA
AATTACTCGTAAATGTGTGTTAAAGTAATAGGATTAAATTAAAATAAACATTTTACGAAATAAGTTCTAGTCAATTATGA
AGTCTTTTCT
>SMED30004304 ID=SMED30004304|Name=Proliferating cell nuclear antigen|organism=Schmidtea mediterranea sexual|type=transcript|length=907bp
CTAAAAGGTAATAATTTAAATTTAATTAGAACATTGTTTTAATATTCGGTTATATTCTCCATTATTCTAAATTATGTTTG
AAGCAAAATTAATAAAAGGCGAAGTTTGGAAAAAGGTTATAGAAGCTATAAAAGACTTAGTACAAGAAGCAACATTGGAT
TGCTCAGAAAATGGAATTAGTCTTCAAGCCATGGATACATCTCATGTGTCCTTAGTTAGTATGCTTCTTCGAAGTGATGG
TTTTGAGACTTATCGATGTGATAGAAATGTCAATCTCGGTCTTAATATAACAAGTGTTTCAAAAATTTTAAAGGCGCTTG
GTAGTAATGATTCCCTAACAATGAAAGCCGCTGATTCAAGTGAAACAATTTCGCTTTTGATAGAATCTGCAAATGAAAAT
GAATTATCTGAATACGAAATAAAATTAATGGACTTGGATGGAGATCACTTAGGTATACCAGATACTGAATATAAGTGCAT
TGTGAAAATGCCGAGTTCAAAGTTACAAAGAATATGTCGAGATTTAAGCCAAATTGGAGAAGCAATCACGATTTGTGTTA
CTAAAGATGGTGTAACTTTTGCTGCTGCTGGAGATCTAGGAAAAGGAGCTATAAAACTTAATCAAAATTCTAGTGCTGAC
AAAGAAAATGAAGGAGTAACTATTGAAATGACCGAACCTGTTTCGATGACATATTCATTGAGGTATTTCAATATGTTTAC
AAAGGCTGCACCTCTTTCTTCTCAAGTATCTCTGTCGTTGACTGAAAATGTTCCAGCAGTTATTGAGTTCATAATCGAAG
ATTTGGGATTTATTCGATATTACCTAGCTCCTAAAATCGAAGACGACGAGTGAAAGTTTAACTATTAATGTTGTATGTTT
AATAGATGTTATTTTGTTTAAAAAAAA
>SMED30012792 ID=SMED30012792|Name=Netrin 1|organism=Schmidtea mediterranea sexual|type=transcript|length=2046bp
CATCTGCGTGCGAGATCGAGTGCTGACTGATTTGAAGAATACCATTTTTGCAATACAAAAAAACACATGAAAATTCTTTC
TCCGCCACAATATACAACGGACACAAAATAAAGATTCACTTTTTTAATTCATATCAAAAAGAAAAAATCAATATCAAAAC
AAACGAAGTGAAAACCCCACAAGAAAAATGTGCAGATATATTTTCACAGTTTATTTTTCAATGTTTTACAAATTTGTGCT
ATCGCAGTTACACATGAATAATTTGACTCCCAGTCCATGCTACGCTCACGGATCCGCATATCAGTGCATGCCTCCATTCA
CAAACATTGTAGAGAATATGATTCCAAAGATCGAAACCCTATGTAAAAAGAAGGAAGTCTGTCCTATAAAATCTCCAAGG
GATAATTTAATCACAGATTATCACTTGACTCACAATCAAACATGCTGGAAGTCAGTTCCGCTACAAGCTAATCATGTAAA
TATTACGTTTTCATTTGCTAAGAGATTTGAAGTTTATTACATCTCTTTGACACCATGCGAATCAGGAATTCCCAACGCCA
TCGCCTTTTATAAAAGCAACAACTTTGGAATCACTTGGAAACCTTGGCATTATCTTGCAACAAACTGTTTATCCATGTTC
AATATGAAAGAAACGACAATTCCCAGTCTACAGTTCAATGATTTTCGAATTCAACGTAGAGAACAAGCTGCTAGATGTTT
TCCATTAAAATTGTCGCTAACCGACAACGAGCCAGTTGTGGCTTTCAATACAGTTTTAGAAGGCGCCTATGAAAACGTTG
GGGATTCCAGCTTGATTGACTGGATGAGTGCGTCTGATATTCGGATTACATTGTTCAAATTTGACAATCCTAAACCGAAA
AAGTCGAATATTTTCCGATTCGCTAGAAATTCTCCGAAGAAAATTTTGTCAATTTCCGACATTTCAATAGGTGGACGATG
CAAATGCAATGGTCATGCAAATGCTTGCAATAAAAATCCTATAACGAATAAAATTGAATGCGTTTGTCAGCATAACACTG
AAGGACAAGATTGCGAAAAATGCAAAAAAAACTATTTGGATCTTCCTTGGGCAAGAGCCACAGTAAACTCTCCAGCTCAG
TGTAAAAAATGTGAATGCAATTTGCACTCTTCAAACTGCTATTTCAGTCAAGATCTTTACATTCTTCGAAAGCAAATCAC
TGGAGGCGTATGTACAAACTGTCAACACAATACAGCGGGAGTAAGATGCCATTATTGCAATGATGGATACTACAGAGATT
GGACAAAGCCATTAGATCATAAATTCGTTTGTAAAAAGTGTACTTGCCATATCATTGGGGCTAAATTTCCGGATAGATGT
GATAAAAGAACAGGACAGTGCTTGTGTAAAAGTGGTGTAACGGGAGAAATGTGTAATAGATGCGCATCAGGATTTAGACA
AAGTAACTCAACAGAAACCCCATGCGTAAAATATATTCAAATGGAGTCAGCTGAAAACAAATGCAAAGCGTGTGATTCGA
AAAGAACTAGAATTAGATTTCAAAAATTCTGTCGCAAAGACGCAGTGTTTTCTGCGACTTTACAATCTCAAGAAATCATT
GGGGATTTTATTCGGTTTGATGTCACAGTTGATAATATCTGGCATAGTAAAAATTCAATCAAAGATTTATATCCTGGAAT
ATTTAGCCAGCCCATAATGCGCCCTTCAAGCAAGTCCAGACTAAATAACAGGCCTACAAATCCGATATGGCTGAAATTGA
GGGACCTGAAATGCAACTGTGTCCAGTTGCAACTGGGGCAAACCTATTTATTAATTATCAAATCAACGACTTATCACTAT
CAAAATCGTGCGGAGCTTCTCATCAAATCTCCTCGAAGCATCATTTTAACATGGAATCCGACATGGAAACGACGACTCAA
TAGATTTAAACAAAAAAAAGAAAAAGGCGAATGCTCAAAATTTCAAAATCGTTCTCCGTTTCGAAAACCGGTGAACTGGT
CAACGCCGAATTACAGAACTGACTCAAATAATTTGTAAAGTTTCTC
>SMED30014446 ID=SMED30014446|Name=SMEDWI-2|organism=Schmidtea mediterranea sexual|type=transcript|length=4773bp
ATTTAATTTTCGAAAAAAAATATGGAGGAAATACCAGTAAAAGTAGCTAAACTTGAAGCGAATGGAAGTGAAACAAGAGG
TAAAATGGGAAATCGTGGTGGCCTGAGAAGAACATTTAGAATTGTTGAACCTAACTTGCAACCTGCTGATATCACTGAAA
AAACAGGAAACATAGGAAGAACAGTTAAATTACAATCAAATTTTACCAAATTCTCTGTTTTGCGTTCAGAAAAGTTCATG
ATGTATGATTCCGTGTTTTCTCAAGTTGGTTTGTCACCCAAAGTAAAATTACAGTTTATTGTTCGAGTCTGCCAAGAAAA
TAAATTGCCAGGGGCTTTTTGTTTTGATGGAAGACGTCTATTTACTAGTGAAAAATGGCATGGAGAATCGGACATTCAAG
AATTTAGTCATGATGAAAAAAAAATGACACTAAGATTGGTATCAACTATCCTTCCTGAAACTGAAGAATATTACCAAATG
ATCAATGTATTACTCAATAATTTACAAGTAATGTTAGGTCAAGAAAGAATTGGTAAAGGTTATTTTTTAAGCCCAAATAT
TACAAAAGATGAAGTTCCCCGTAATTCTGGACCGACATTTTTTGAAACCAATAGTTTTAAAGTTCTTGGGGGCTTTGGTA
CAACTTTACAAAGAGGCACAAAACCAACTGGTGAATTAACCACTCTTTTATATATTGAGCGTATTAACAGAGTATTAAAC
GATAACTCTGTTATGAAGGCATACAACAGAAGAAGTATCGATCAGTTGATAGGCAGAGATATCATAACAAAATATAATAA
CAAAACTTATAGGATAAGTGAAATAAAAGAAATGAACGTAGATGAGAAATTTGAAATGGGTGGTAGAACTTTATCGTATG
CCGAATATTTTAAGGAACGATATAATATTCGATTAACTCAAGGCGATCAACCATTTGTTCTAACGCGAGTGAAGAAACCA
ATGCGTCGAGAAAGAAAGAAAAAAGACGAAGAAGGAGTGGAAAAAGAGAAAGAAAAAGAACGCGAAAAAGAGGCTCCAGA
AGAGAAGGACATGACTCTCAATATTCCTGGAGAGTTGTGTTTCTTATGTGGGTTTTCAGATCAAGAGAAATCGAATATGG
ATCTGCAGAAGAATTTAGGGTGCGTTTTAAAAAGAGAGCCACGTGAAAGATTGGATGATATACCAGCTTATTGTAACTGG
ATTAAAAATAGTGATGCGGCAACTGGAATGTGCAATAAATGGCAGCTGAAAATAGATAACAAACCATTAGAAATCGAAGG
TAGAGAATTACCTCCATGCGATGTAATAAGTGGTGGCAGCAAAATTAATGAAAAAATGGGAGATGATTGGAAATTTGGTC
GAGTTCAATTTGATATTAAGCGTGATCGTAAGCATGAAATCGATGTGGTCATCGTTGATCGAAATGACTTTCAGTATAAA
AATTTTATGAATGATGTGGAACAAGAATTAAGAAACATGAGAATTGATGCTCGAGTTGGAAAGGTTAATACTTGTGGTCC
AAATGATGTTGAAAGATGTTTGAATGATGCCGCTAGAAGTGGAAGTGGGTGTGCTAAAATGGCGCTGGTATTCGTCCCAG
ATGATAGAGTATATGCCAAGGTGAAATCATTTACCATGAGTACTGGTCTTTTAACCCAATGTGTAACAACACGTAATGGT
ACTAATAGAAACGATAAACGAAGAAAGGTGGTTTCAAGTAAAACCGTGATGCAAATATTTGCAAAATTCGGATATGATCC
ATGGACAGTTGAAATTAAATTGAGACCAACTATGATTGTCGGTATGGATACCTATCATAACAAGTCCAGCAAGAAATCAA
TTCAAGCATCTGTATTTTCGATCAACAGCACATTTACACAATATATGAGCTTTGTTAATTCTCCTAAAGGACGTCAGGAA
TTCCACGAAACTTTAGGCAAAAATTTCAATTTAGCGTTAGAAGATTTCAAGAAACGTTATGACATATTGCCCCAGCGAAT
TTTAGTCTTTAGAGATGGTGTTGGAGATAACCAACTTCAATTTACTAAAAATTTTGAAGTTGATGCAATGAAACCGCTTA
TTGAAAATATATACAAAGGCAACGGATTCCCTGTTCCACAAATCATTTATGTCATCGTAAAGAAAAGAGTCGGCACTAAG
CTTTTCAACAGAGGAAATAATCCAAATCCTGGAACTGTTGTGGATAAAGAAATTGTCAAACCGAATTTCTATGAATTTTT
CCTAGTTTCACAAAGAACAACGAAAGGAACTGCAACTCCAACTAATTATAATGTTTTAGAAGATACTCGATTGCTAACGA
AAAAAGGTACTATGGATCCTATGGCACCGAATGAGCTGCAAAAGATTACCTATGCTCTAACTCATTTATATTTCAATTGG
ATGGGAACTATCCGAGTTCCGGTTCCAGTGCATTATGCGCATAGATTGGCTGAATTAGTTGGAAAGGTGCATAAGGGCTC
TAATCCCGTTGCCATTAATGAAAGAATAAGAAATAGGCTCTTCTATTTATAAAAAGATCAAAACTTACATTTTTCTGTGT
TGATTACTATCCCAATTGTGGATTTAATTGTTTCTGAAAGCTGTTCATTCAAATTCTTCTGAAATCCATATTGATTCGTA
AGCAGCCTGTATTTGAATACTAAAATTGATAAAGTAGTTATAAGTATTGTAAAAGTATTAAATATTTGTTTATGTTATTC
GTAATACACTTGGGTTATTGCAATGTTTTAGTTGTTTTTCAATGATAATAAAGCTTAGCTGTATAAATTCTTTTAGGTTT
TTTAGAAGGTTTATTTATATCGTGTAATTGATTGTCGTGTTAATATTAATTAAATAAAAGTTTGATGTAATTGCTTTGTG
TTACGCATAATAAATCTTACGAAATTTCTGAATACAACTGCTGGTGATCGGGATATTGAATGCCGGGTCGTTTGAATTAA
TAATACAAATGCTTTCGATTATTGTCATTAATATTTTCTCCAGTGCGTAATGGGCCAGAGTTCCGGTGCCATGATTAATG
GTTCCTAAATTTTAGAAATAACATTTTGAATGACATGTTAAATGTGTATATAATAAATTTTCTAGTTTTTTTGCTAAGAT
TTTGAGAAATTTTAGTTACCTGTTTTTGTTAATTGCTTTGATAATTTCAATGCTGACACGTGCAGAATTTCACACATCTC
CAGCAGTCGACTGAATTCTTGGATTTTGTTTGATACAATTATGCTTGCAGATGTTATGGCGCATAGATCCAAGAAGGCTG
TGAATCTTGGGTCTTGTTTTGTAAAGGAAGCTTTGTAAAATATGATGGGACAGTTGCACATAAATTCTAGGAATCCAGAT
TTTAGGCAAGATTGAAGTAATGTTGAATGGTTCTATTATAGAATAATCAGTTGCACTTTTAGATAACCAATATACCACAA
AGAGCCATCCGCAAATTTTGAAAGCCCAAAGCATTGAATCGTTGTCTATGTTACTCAGTATAGACAGCAGTAAAATGGCG
CTTTTTTGATATTGTAAACCGAAATGATTACTAGAATTCATTGTTTCGTCAAGAGATTTCGCGAAACTCTGAATATTATC
TGATTGATTTGATACTATTCCTTGGAGTTTACAAATTGATTTCAGTTCCAAATTCATACTATTGTGTTCTGCTGATTTGC
TACTATTGGAGCTCTATAAGGAATTATTAGAATCAATTTTGGTTTGATAGTGGGTTTTGCAATCATTGGCTTATTGTGGG
TTGTGTAACCATGGAAGTAATTGTGAATAGTTATAGTGCTCGCTCTGCATAAAATAATATTTAATCAGTGCTTTAAAGTA
GATTTTTAACGATTATTATGAAAGATTTCTCTTATATATTAATGTTTTTGTATCCTTCAAAAGTTTACTAAAAATAATGC
ACATTTGAAATTTACGCGTTACCATAAAATGTCTGAAGTGGTCCAAAGCAATAAAAGCTTAAAAAGAAAGTTCTTGTTTT
TAAATGTATTGGGATATACTCCCACTAACCTTCCGTACTCCGCTGCCTCCCTTCCCATCACATACAATATTTGCTAGTTC
TACTTACAGAGTTGAAAATCGAAAAATTTTCCCAGTCAATGTCCATGATTATGCTGAATAAATTTTGTCCGAACATAATA
GAGATTGCCTCAGCCAGTATTTCTTTCGTTGTTGAGTTTTGATGAATGACATGTGAGACTAAATTTATGGACAACCATTG
ATTTTGGTTTTGCTGAAGTTGAGACTTCTTACGGTAAAGATAAACACGAAATAATGATAAACTCTAATCAAAAGATATTA
CAAACAAATATTATGATATCAAAACCTTCTGTCGAACCAAGGCACTTGTGTGAGATAATAAATGAAGAATGAATTCGAAA
AAGAAGTCATAATTCGAACATCGCTCCAGAATGCTGTCACTGACTTTGACTTCAATCACATTTTCCAATATTGAAATCAG
ACCGGCGATCACATGGACCCTTGATGATTCAGACTGACCACTGATTACGGGTTGATCGGTTAAATTATCTAGAGATTAAA
ATAATAGTCGGTAAAGGAAATTATCCATATAAACGGGAAACAGAAAAGTTTTAACCATGGTTTCAAAAGCTTTTGGGACT
ACCCCCAGCTTTGCAAAACAATTTTTTAACGCTCTTGTTATGACTTGAATCAACATTGTACAATTGTTACAATAGACTTG
ATGATTTACTTATTTACACCCCATCTAAATTGTTTCTTAACCCTTTGCCACCC
>SMED30027701 ID=SMED30027701|Name=ATP dependent RNA helicase DDX5|organism=Schmidtea mediterranea sexual|type=transcript|length=2292bp
ATAATAATAATTGTTGTTATTATTTGATGAAACAATGAAGAATAGTGTCGATGTTTACTAGACATACAAGCCGTTACAAC
AAAGCGGCACATTTACGGGACTAAGCTGACATGTCCATAAATGGTCTATGTTAGCCGTGTGGGCGCATATCTGTGTGCAT
CAACACGGTAATTTCTCACACGAGCCGGTGGAGTGGTCTCGTTCATCGTGTTGCTGATAACCACCGAATGATGGTTTGCT
GTTGCCAAATAGCTCATCAGCCTCAGAGATTTGTGTCGTCGACCGGGGTGGTCGATGGAGCCACAGGTTTTGTCACTGGT
TTCCCACCAAGAAAGCCCGTGACACGACATCGCTTCGTGGTTCTGGTGATCACTCGGCGCGACTTCAATGACACACAGAA
AGACACATAGACAACTGTCCGGTCTCTTATGGGAGTACGTGGATATTGATGATATTGAATATGTCATCAATTATGATTAT
CCGAGCAAGTTAGAGGATTATATACATAGAATTGGCAGAACAGCTAGAAGCGGCAAAAAGGGAATCGCGTATACATTTTT
CAATCCCATGAAAGATAAGCCTAGACAAGCACAAGATCTAATTATGATTCTGGAAGAAGCAAAGCAAGAAGTTCCAACGG
AATTAAGAGACGTTGCTTCAAATCGAATACCTAGCAAACGATTTAAAAGTAAAAGAAATCGATCTCGTAGTAAACAGCGA
TCAAATAGTCCTAAAAGGTCAAGAAGTCGAAGAAGTAGAAGAAGATCAAAGAGTTCCAAAAAATCGAGAAGTCGTAGTCG
TAGACGATCTCGCAGTCCAAAACGTCGATCGAGAAGTAATCGTAGAAGGCGCTCACGTAGTGTAAGAAGAAGATCTCGGA
GTATTCGAAGATCAAGAAGTAGAAGACGTCGATCTAGAAGTAATCCAAGAAGATCAATAAATAGACGTCGAAGAGCAGAA
AGTAGAAGAAGAAGCAGAAGTCGGAATGATAGAAGATTGGCTGTTCCTAGACGACGACGCGTTTCACCTACTCCACCAAG
GTTACGTAGAAGAAAAAGCAGTTCCAGCAGTATATCAGGAAGTCAGTCTAGTGATTCTCGTTCCAATAGTAGCGTCTCAA
TAAAACGGGAAGCACTAGCCGATAAGATTCCAAATATAGCCAATGAAAAAGAAACTCATCAAAATGAACTGGAATCAAAA
CTTCATGACGAATTGAGTTCGGAAAATAGAGAGCCATTGGATATCCAATTGACAGTTAATGAAAATGAATTTGAAATCGA
AATGTCGAAATCAAGATTGAAAAAAGATATTGGGGAAGAGAGAAATAAAGAAAACAAAGTATACGAAAATTGCATGTGGC
AAAATAACTTTGCGGAAGAGGTTTTTGAAGTGCCTCCCGTTGTTGCACACCACGAGGTCGATATGGATATTGATGAATAT
CGAAAGTCATTTGATGCATTGAATCAAACAAGGCAAAAGGAGAAATTATTCGATTCTATTCGGAAAATAATAAAAATCGA
TGACGCTGCAAAACAGCGACCGATAGAAGTTATGACTAATGAAGAAATCGATATTATAACAACTGTTGAGCAGCCACCTG
TTAAAATGACGGCAGAGAAATCTGTCCAATACGAAGAACCGGAATTGGATAATATTGCAATGAAGGAATCAATCAAATCA
GATGTTGAAGTACTTTTGGAAAAATCCACTGAAAAGGATCAAAAAGTCAAAGTTCTGGAGTCTGACGAGGAACCTAAAAC
CTCCAATAAATCTAATTTGGATGAAAAATGTAAAAGTCGAAATAAATCCGATAATCAACATAGTTCAAGAGATGATCGTG
ATGAGAAATTTAAAAAAAGCAAAAATGAAAGCGATAAATCCAAGCGCACCGAAGACAAAAGCGATTCTAGAGTGGATAAT
AGAAAAGAGAGCAAACGTGACTCGAAAGGTGAACCACGTGTTTCTCGTTGGGAGATTCATTCATCGAATAAAAATGAAGA
AATCAAAGCGATGGGAAAGTGGGAAACTAAATCTGAGCTTAATCAAGATAAATGTGAAGCTGAAAAAACGTTTGATAGTA
AAGAGGAAAAACCTTGTAATATGCTCGAAGAAAAAAAGTTGGAAAAGAAGCATGAGCCGAAAGATGACAAGAAGCGGGAT
CATAAAGATGAAAAGAAACACGAACAGAGAGAAGATAAAAAACGTGATCATAAAGAGGATAAAAAACACGAAACGAAAGA
CGATAAGAAACATGAAGTGAAAGAAGAAAAGAAACATGAAACGAAAGACGAC
>SMED30022921 ID=SMED30022921|Name=Netrin 1|organism=Schmidtea mediterranea sexual|type=transcript|length=1370bp
GATTTTTATAAGAACAATAGCAATTCTAAATTTATCAGTGTGGATTCATTCTGTCAAGGTTATTTTTGAATCTCAGTCTT
CACCATGCTATTTAAACGGGAGGCCTCAACAGTGTTTGCCGTCTTTTACAAATATTATTGAAAACGTTAAAGCAGAAACT
ACAAGTACGTGTGGGACCAATGGACCGGAACAGATTTGTCGGAATTGGTATGACAAGACAACTGCAGAAGCCCGACAATA
TTGTAGTATATGCGACGCAAAACATCGTTCATATCCACCATCCCATATAACAGACCGACACATTCCAAAAAATCAAACAT
GTTGGTTTTCTGGACCGTTACAGGAAAATCCTGGTCAAAATGAAGTCAATATTACTGTTTCGCTGAAGAAAAGCTTCGAA
GTTTATTACATTGCATTGCAAATCTGCGGAACACTTCCAGATTCAATAGCAATTTACAAAAGCTTAGATTTTGGAGTTTC
ATGGAAACCTTGGCAGTTTTTTTCACAAGATTGCTACAGGGCATTTAAAATGCCAACAACCAACGAACAGAACTCTCAAA
TCACTCCTGCAAACATTCACGAAGTTCTTTGTGTTGAATTAAAAGCCCCTGAACGATACAATGATTATGGACAAGCAGAA
ACTGTGTTACCGTTTAGTACCATTTATGGGCGACCATCTGGACCTCCATGGAGTCAAGATTTAGTTGAATGGATGACAAT
GACAGATTTGAGAATAAGTTTGATGAGATTTACACCGGAAAAATCGACTCCATACAGATTGAAAGAATTGTATTATCATA
CCGAATCACTTCCCCAAGCACCACCGAATTTAGTCAACCATAAAACTGAGCCGATTGTTCATTTTGGTTTGTCAGATCTT
GCTGTTGGTGGACGCTGCAAATGCAATGGCCATGCTAATCGATGTTTGATTGATGATAACGATGAAATCAAGTGTGACTG
TCACCACAACACCGAAGGTCAAGAGTGTGAAAGATGCAAATCTAATTATCTTGATCGACCATGGCAAAGAGCAACCAGGC
AAAATGCCAATGTCTGCAAATTATGCCAATGTTACAACCATTCTGATAAATGCATGTTCAGCATGGGCATGTACAAACAA
ACTCGAGGACAACATGGAGGGGTTTGTTTGGAATGCAAACACAATACAGAGGGTCACGCTTGTGATGAATGTGCTGCATT
TCATTATCGTGATATAACAAAACCAGTTACACATCCTCAAGCTTGTACAAATTCATGGCACAGGTAGGTGAAGGCGCAAA
GCACCCGTTAATACTATTAATTGCGCACCGTGAACTAACCAATAAAAATCGAGTCAGGTTGCAAATGTCACACTATAGGA
TCTATTAAAC
>SMED30001900 ID=SMED30001900|Name=Bruno-like protein|organism=Schmidtea mediterranea sexual|type=transcript|length=1713bp
CTTTTGAGGGAAAAATAATTCTTTTATTAATAATGTTTCATCATTGGAATATAAAATAAAATATTAGGAGAAAACATAAG
TTAACCAACAATATTAGCCACATATTTCGTTTAATTATTGGTCTATCACTTTTCATTTTAACCAATTGTCTTGAGATAAT
GGAGAGTTTATCCTTAATTTTTATTATAAATCACTGGTTTTGATAGACTTATTAACAATGGAAAATTATTTCAAACTTTT
CGTTGGTCAGATACCTCGAAATACACCAGATAAAGAAATTAGACTTATTTTTGAAGCTTTTGGACCAATCTTTGAATTTT
TAATTTTAAGGGACAAGAAAACTGGTTTTCATAAAGGATGTGCTTTTGTTACATTTTATCATAAAGACTCTGCAGAAAAT
TGTCAAAAGGCGTTACATGACAAGAAAATCCTAAACGGGATGAATCGACCACTTCAAGTGAAACCAGCTAAAAATAAGTT
AGCAAAAAATTCAAGTTTGAATGGGGAAAATCAGACAGAAGAGCAGGTTTTCAGTGATGAAAGGAAATTGTTTGTTGGAA
TGTTGAGTAAAAATCAAACTGATGAAAATGTACAAAATATGTTCACAAAATTTGGCAAAATCGAAGAGTGCACTGTATTG
AAAGACCAAAATGGAAATAGTAAAGGATGTGCCTTTGTCAAATTCGTGAACCACACCGATGCTTGTGCAGCAATAAACGC
TCTTCATGCCAGTCAAAAAATGGAGGGTGCTTCATCCAGCCTTGTGGTTAAATTTGCTGACACGGATAAACAAAAACAAA
TAAGAAAATTGCAGCAATATATTCCAGATTTAAGCATTTTAAATTCGCGCATTCCTATTAATATTCCATACTATTCACAA
TACGGTACAATAATTAATCCAGAATATAATTCAACAATTCCTTCAAACCAATATATTCGAAATTGTAACGAATATATTCT
TGCTGCTCAAATTCAACAAATAAATTTGTTTTCTAATTTATCATCGGTCTACCCATCTAATTCAATAGTGTCAATGCCAT
CCAGCCATCCGGTACCAATGATTGACTATTCTGTTCCTATTAACAACTTTCCACAAACTGACCAAATAACAATGAAACCA
ATTGTTGATATACAGCCAATACCTCCAGAAGCTGTACACCGAACCATCGTAACTCCAGGATCACTCTTATCATATCCTTT
TCTTAATGTTCAAATTCCTACAGTATATCCAAACGCCTTTTCCAATGGAAATTTAACAAGTACAACACAAAGTTCAACAG
ATGTGAAAACCACTCCTTTGCCAACAGGTCCTGAAGGCTGCAATCTATTTATTTATCATCTGCCACAAGAAATTGGTGAT
TTACAATTATATCAAATTTTTATGCATTTTGGAAACGTTATCAGTTCTAAAGTTTATGTTGACAGAGCAACAAATCAAAG
CAAATGTTTTGGATTTGTCAGCTACGATGATCCGGCATGTGCCAATGCCGCCATCAAATCTATGAATGGATATCATATCG
GAACAAAAAGATTAAAAGTTCAGTTAAAAAAACCAAAAGAAGCAATAAACGAAATGAAAAACTTCAATCAAATGTTATCA
TCAACTCCCGACATAAACCAAGTATTTTGTTCATCCTAATGATTGTGATATTTTACAGGATTTTCTCGACCATATTGAAA
GCCTGATTGGTGATATGTAAAATGACCCACCGT
>SMED30020435 ID=SMED30020435|Name=Myosin heavy chain|organism=Schmidtea mediterranea sexual|type=transcript|length=3615bp
TCTGAATTTAAACAAAGATATTCAATTTTGGCCGCAGATGTAATAGGGCAAGGATTTATTGATGGAAAACAGTGTACTGC
AAAGATAATGGAAGCAATCCAATTAGATAAAAACAACTACAGGCTTGGATCAACTAAAATCTTTTTCAAGGCCGGAGTAC
TTGCTCAGTTAGAAGACATGCGAGATGAGAAATTATCATCCATTTTGTCTGTATTCCAAGCACAGATGAGAGGATATATA
ATGAGAAAAGCATATAAGACATTGCAAGATCAACGAATTGCTCTTTCTCTCATTCAAAGAAACATCAGAAAATATTTATT
ACTAAAAACATGGCCATGGTGGAGACTATACACTAAAGTCAAACCACTCCTCAATATCGCAAGACAAGAAGAGGAGATGA
AGAAAGCTGCTGAAGAGTTGATTAAATTGAAAGAAGAATATGAAAAGATAGAAAAATTTAAAAAGGAATTGGAAGGTCAA
AATGTGGTTTTGCTTGAGCAGAAAAATGATTTATTTTTACAACTGCAAACAGAACAAGACAATCTTAGTGACGCTGAAGA
AAAGATTCAAAAGATGATCAATCAAAGAGTTGAAATGGAGAACCAATTTAAAGAATTAGAAGATAAATTAATGGATTCGG
AAGACATTCAAGATGAATTGGAAATCGAAAAAAAGAAATTGTTGTTGGATATGGATGAATTGAAATCGGACATTGACGAA
CTTGAAGTATCTTTAGAAAAGTCTGAACAAGAAAAACAAGCTCGCGATCAACAAATTAATAATTTGAAAGAAGAAATTCA
AAAACAAGATACTTCTATCGGAAAAGTTCAAAAGGAGAAAAAGACTCTAGAAGATAACTTAAATCAGACCAGAGAAGCTT
TAAATGCTGAAGAAGACAAGGTCAATCAGTTGACTAAAATCAAATCAAAATTGGAATCTGAAATAGATGAAATGGAAGAA
GCTCTAGAACGGGAAAAGAAGATCCGTGGAGATGTTGAAAAACTTAAACGGAAATTAGAAGCTGATCTCAAATCAAGCCT
TGATAACTGTAATGAGTTGGAAAATATAAAGCAGGAGTTAGAAGAAAATATTCAGAGAAAGGACATTGAAATATCAAAAT
TAAATGTCAAATTGGAAGATGAATCTTCAATTGTCACTCAACTTCAACGAAAAATTAAGGAATTACAAACTGTTATACAA
GATCTTGAAGAAGATCTAGAAGGTGAGAGACAAGGTCGATCAAAATCTGAAAAAGCTCGACAGTTGGCTGAACAAGAACT
CGATGAACTAGCTGACAGACTTGAAGAGCAGGGAGGTGCTACTATGGCACAAGTGGAATTGGCCAAGAAACGAGAAACTG
AATTAATGAAACTTAAAAAAGAATTACAAGATACCAATTTACAGAATGAGCAAATACTCAATGGTTTGAAAAAGAAAAAC
CAAGAAATTACCAATGAATTCTCCGAGCAAGTCGATCAAATAAATAAATCTAAATCAAAAATCGAAAAAGAAAAGGCGAC
TCTTAAGCATGATCTGGATGAATTTGTTTCTCAATTAGAATCTGTTTCAAAGCTAAAGGCCAATACAGAAAAACATTCAA
AACATTTAGAAAGTCAAATTATGGAATTGGAAAACAAAATAGACGAACAGAATCATCAAATATCAGAAATGGTTTCACAG
TCTAATAAAGACTCTCAAGCAATAATGTCTTTAGAACAACAACTAGAACAAGCAGAAAATGAAAATCACCAATTGAGCAA
ATCCAAACAACAATTCCTCCAACAAATTAATGACTGTAAATCGACTCTTGAAGACGAAGAAAAAGTAAAGGCCAAACTTT
TCAATGAAATTCGTAATCTGAATGATGAAATGGGCAATTTAAGATCCAGTTTGGAAGAAGAGCAAGAATCCAAAAGTGAA
ATTCAAAGGTCACTAGCAAAATCCCAATCTGAACTTGTACAAATGAAAAATAAATTTGATAGTGAAGGTTCAGGTCGCAT
TGAAGAAGTTGAAGAAGCTAAACGTAAGTTGCAAACGAAACTTCAAGACATGGAACAAGAATTTGACGCTATTAAAAATA
AATGCGCGCAACTTGAAAAAATCAAAATTCGTCTCACTGGAGAAATCGAAGACATGAGTGTTGATGTTGACAGAGCTAAT
GCAGTGGCTAATCAACTAGAAAAGAAACAAAAGAACTTTGACAAAACCGTCAGTGAATGGCAGTCTAAATGCAACTGCCT
TCAAACTGAAGTTGATGTTGCAAATCAAGAATCAAGATTTTTGTCAACCGAAGTATTTAAATGTAAAAGCCAAAACGAAG
AATTGCTTTCACAAATCGAAAATCTTAAACGAGAAAATAAGTCTCTCAGTGATGAAATTCACGATTTAACGGAACAACTA
AGTGATGGCGGAAAATCTGTTCACGATATAGAAAAGGCAAAACGAAAATTGGAAATAGAAAAGGAAGAACTGGAACAAGC
TTTGGAAGAAGCTGAATCAACATTGGAAAATGAAGAAGCCAAATTACAAAGAGCTCAGTTAGAGTTGAGTCAGATGAAAT
CAGATGTAGAAAAACGATTGTGTGAAAAGGAGGAAGATTTTGAAGGAACTCGCCGTAATCATCAAAGAGCAATGGAATCA
ATGCAAGCTTCTTTGGAGGCAGAAATTAAAATTAAACAGGACTCAATTAAAGCCAAGAAGAAGTTAGAACAAGAAATCAG
TGAACTCGAAGCCAATTTAGATCAAATCCAAAGACTGAAATCAGATGCTGATAAGAACAATTCAAGAATGCAACAGCAAA
TTCATGATTTACATTCAGAGATTGAACAAGAACAACACAGTAAAAACGAAGTCCGAGAACATGCTCAGATGGCTGAAAAG
AGATTTTTATCGGCTAATGGAGAACTAGAAGATTTGCGAGCCAAATTTGAACAATCAGATAGAAATCGTAGAAATGCTGA
ATCTGAGCTTCAAGATTTGAGTGATAAATACAATGAAAGCCAAGCACTGATCACTTCACTTCAAAGTTCAAAAAGAAAAA
TGGAAATGGATTACGTAGCTATGCAGTCCGATCTTGATGATATGTCAAACCATGTTCAACAAGCTGAGGAAAGAGCAAAG
AAAGCAGCTGGTGACGCTGCCCATATGTCAGAACAACTTCATCAAGAACAAGACAATAGTTTGCACACGGATAAAATGAG
GAAGCAAATGGAAATTCAAATCAAAGAAATGCAGTGTAAACTAGAAGAAGCCGAAAGTAATGTTTTGTCAGGCGGCAAAA
AGACTTTGGCAAAGTTAGAGTCAAAAATACGAGAATTGGAAACTAATCTTGATAACGAAATGAGAAAACATGCTGAAACT
CAAAAGAATTATCGCAAAGTAGATAGACGAATGAAAGAACTAACATTACAATTGGAAGACGATAGAGCTCAACAAGAAAA
ATCAAATCAAGTTATTGAAAAATTCCAAGGACAAATCAAAACCCTTCGAAAACAATTAGAAGAATCAGAAGAAGTTGCCG
TTTTGAATATTGCCAAATTTAAGAAAGTACAATGTGATTTTGAAGAAGCCGAAGAAAGAGCGAATGAATCGGAACAAGCT
TTACAAAAATTAAGA
>SMED30035063 ID=SMED30035063|Name=SOXP-2|organism=Schmidtea mediterranea sexual|type=transcript|length=2664bp
TGTACATTGAATTCAATATTTAATAGTGATTAACTTATCCAAATTTTCAGCCACTTTTAGTTTAATTTGCGTGTTTTGTT
GAGCAGTTTTTATTGGCGCATTTTATAAAAAATACAAAACCAATCCAAATCAGTTTGAATCATTGTCAGATTTTAGTCGT
GCAAGATACATTATTTCATTTTAATTCTCTAGCATATGACATTTAAACATGGAGCTGAATTACTCGTTGAATGATAATAA
TTTCAATTTACAATTAGAAAGCGAGTCCTTGACTTATTTCGATTTATGCCAAAAATGTTCAGCGTACCAAAATAGTCTTA
GAGACTGGATGAAACATTTAGTTGACAGACATCCTTTACCTATTCAGTGGATGAGTGATTTGGCAAATAATTTAACAGAT
GATAAATTATCTAAGCCATATTCGACCGTTACAGAATCCCGTAAAAAACGCAAGAAGCATATCACTCCAAGGCCATTGAA
TAGTTTCATGATATTTTCCAGACATTTAAGAAGAAATGTTCTTATTTTCTTTAGTGATGCTTCAAATTCAGAAATAAGCA
AACTTCTTGGGAAGTTATGGAATAACATTCCAGCTACCATTAAATCGATGTATGATAATGAAGCTAAAAAGCTTGCAATT
TTACACTCCAAAGAATTTCCTGAATATAAATATCAACCTAAAAAGAAAAAAAAACAATATCTAACGAAATGCCCCATTAA
TTCCACACCTTTGTCGAGTAATTCATATGAAAGTATTCAAGATATTAATTCAGTAACGGATACACTACAGAATTCATCTA
ACAACTTACAAAAGGAAAGTAATCCGACCTCTGTTTCGTCTTATGGAAGCAATCAGGAATGTAATAATTCATTTAAGAAT
CATAGTGTACAGTCCAGCAATTTTCCCAAAGAAATCAACCATAAACCAAGTAGAAAAATTGAAAAGCAATCGTGCAAAAT
AAGTTCCAATCAAATAATGGATGATTCAGAAGGGGCTGAATCCAGTCCATCATCATTTAAAAACACCATTAAAAATGAAA
TTTATTCCTCTTATAGTCCATTTCTAAATGAATCCCATAGCATTCTTTCAGATTTAAGTTACTCTTCGTCTCTCAATAAC
GTTTCAAAACACACTAGCGATTCCATTATAACGAATTCTCCTATCAGAGAAGAAATTGTCTTAGAAAATAATCAGCCCTC
TCAACAGAATAATATAAACAATTACCGACACGTATTTCAGAGAACTTCGACTCTATCCAACAGTTTGTTGACCCCTTTAA
CCATTGTCACAGAAAAAGTTGACGAACAGAAAGATCTTCCACCATCAAGAATCAATATCCGATATGTGAAAATTAACAAA
CTACCGATTCAAACAAAATCCGATTTCCCATTGACTGGATATTTTCAAAATTTCAATAACGACTCAAATCTTCATTTTCA
GAGTTCCATTCACGCCCTGAATACGACATTGACTCCAGATCTGTCGGTAAAACGCGAAGCCACTGTCAACAACAGCATGA
CAGATTATATCAATCGAAAACGTACAAATGTCGATCCACTTATCATCGATGATATATTTGAAAACGAACAATTTGAAGAT
GAGGAAGTGTTGAATTTTGACAATCCTGATAACAATAACAGTTCTCCTTGGTCGTTTAAATCGCCTAACTTTAAAATATC
TACCGGTTCCTCGACACTGTCAGACAATCCGTACAAAGAACAGTTTTGTTCTTCCCCGTTCCATCTATCAGAAACTGATA
CTCAACCGATTCCAGTTCAACGATTGCCAGCAATCGGCTCATGGTTAAAGTCAAACTATTCTGAATTCATGATCTGATCC
AATCTATTATTCTATTCAATTGTCTGTTTTCATACAATTAGGATTACTTGTTATGAATTCTAGCTGATTTTGAGTGTGTT
TATAAAGCTTTTGTAACGCCTTTTCTATACAGATGTTCGATATATATATATAAATAAATAAATATAAATATATAACAATT
TTATTGACATTGTACAGTATTCCATAAGCCCGATCTTGTAACGCATTTATTACTATACTCAATAGACCAATCTATATGTT
CTCATAATTTCTATGTTGTTATTGTACACGGGAATTATTACTATAAATGATGCGAATTTATCACTTTGTAATTGAGGGAA
ATTATCGATCAATTTGCGCTTTCAGACATTTTTCATGTATAAACTTGTTTTTCTCTTTTTTATCATCAAAAATTTAATTT
TTTTGATATCGCTATTCATCATATTATTATTCTCCGTTGCATTTGTTAGATTATGTAAATTTTTATATGAAAATGTTGTT
ATTTATTGTATTTTTATACGATTTTTTGCTTTGGATACACTTTTATCAAATCATATTGCCAGTGTGAGCATTTATTGGAC
TTGTTGTTATTGCTGCTGCTCGAATAATTGCTTCATACGGGGAGTCTCGATATTTTCATGAAGCGATTTTCATGGTTGCC
GCTCTCCGCCGCCGCCATTCGGTGTAAATATCAATTGCATTGCCAGCACGTTTTCTCAAACTCAATTTTTGTACTCAATT
TCAATTGATTTATGTTCATTTTATTTTTCATAGAAATGAGTAAATAAATTTTCATAAATAGTTTTGTTTTCAATATTTTT
AGGAAATTTGAATGTGTCTCTAGA
>SMED30017650 ID=SMED30017650|Name=Cathepsin L|organism=Schmidtea mediterranea sexual|type=transcript|length=1091bp
GATGGGTCCTATCTTGATATGTTTATTATTAGCAGTGATTTCAGAAGCCAGAACCACTTATAGCCATCACAAATCAGTTC
CAAGTGATGACATTTCTAGTAAATTTGAACTAGAAAATATGGTATCTCTCTTGTCCCGTTACGATGGAGAATTCCAAACC
TTCAAACAGCGATATAATAAATCTTATAGTTCCATGCATGAAGAGAATTCCAGATTTTTAAATTTTATATCATCTCTACT
TGAGATCAATCATCACAATCAACTTTATGAAAACGGTCAGAAATCATACGAGATTGGAATCAACCATTTGAGTGATTGGA
GTGGCGAAGAACTCGCTCAACTGACTAAATTGAAACAATCAAACCTGACGGGCTCATCATATCTATCATCCAATGTGAAA
ATTGACCTTCCAGATTCATTCGACTGGAGAGAGAAAGGTGCAGTTACTGAAGTAAAAAACCAAGGATTTTGTGGATCATG
TTATGCATTTGCTACTACTGGCACCCTAGAAGCCCAAATCTGGAGGAAGACTGGGAAATTGATCAGTTTATCAGAACAAC
AAATTGTTGATTGCGATACTAATGATGATGGTTGTGATGGCGGATTTGTGCAAGACGCGTTATCATACATCAAGTCAAAT
GGAGGCATTGAAAATGAGGAAAGTTACCCTTACATATCAAGTGAAACATTTAAACCTAATTCTAAATGCAATTTCTTCAA
AAGCAAAGTAGTGGCCACCGACACTGGATTCGTTCAAATGCCAGCAAATGACGAAACAAAAATGAAAGAAGCTCTAGTTA
CAATTGGGCCTATTTCAGTAGCAATATCTGCAACCAAATCGCTCAAAAGATATACAAGTGGCATTTTTCATGAGGATGAA
TGTGACAAACTAAAACTTAATCACGCTGTTCTGATAGTCGGATATGGAACAGATCAAACTTCCGGGACACCATATTGGAT
AATTAAAAATAGTTGGAGCTCATTTTGGGGAGAAAATGGATATTTCCGGTTACCCAGAAATATAAATGCTTGCAAAGTAG
CGATGATGCCAGTTTATCCTTTGGTTTGAATTGATAAAATTTTGTTCACAC
>SMED30019754 ID=SMED30019754|Name=Cathepsin L|organism=Schmidtea mediterranea sexual|type=transcript|length=1366bp
ATTTAATTTGCTCTATTTTATTTATCAGTAATTTGTTTATTCAAAATATATAATTAACATTGTTTTGGACATCTGTTATG
AAGTTAGTAATAGTTTGTGTTATTCTCTGTTCAGGCCTTGTTAATTGTAATCCAATAACTAAGCTTTCTGACAATGAAGA
CAATTTAATTTTTGATTCAAACCTTTCTAATTCCAAGTCAAATCATGATCATTTTATTGTCAACTTTGGTCAACTAATTA
CTTATGATTCCAAGTTAGTAAAAGAATTGTCGAATTTATTAAATGAATTTCTACAACTCCCAACAATTGAGTTTATAAAT
CAAATAAAAATAGAATGGACAAATTTTAAAGTTAAATTTAAGAAAATTTACAAAGATGTTAAAGAAGAATCTGAACGGTT
TTTGATATTCTTAAAAAATTATCTCAAAATAAAAAGCCACAATTTAAAATTTAATTCCGGAACAGTTCAATTCAATATGT
CAACTAATCATTTATCTGATCTAACTGATTCTGAATATTCAACATTATTGGGCTTTAAATTTGGAAAGCGTAAAGATGGG
CCTTCGTATTTGTCACCTATGTCTCGCGTAATGTTACCGAACCAAGTTGACTGGAGATTAACTGCAGTTACTCCAGTGAA
AGACCAAGGAAATTGTGGGTCATGTTGGGCTTTTTCTGCAACAGGCTCGTTAGAAGGTCAACATTATAGGAAAACGAAAA
AACTTATCAGTTTATCTGAACAGCAATTGGTGGATTGCAGTCTAGATAATAGCGGTTGCAATGGTGGGTTAATGGATCTG
GCTTTCACATATGTCAATGATAATGGGGGTATTGACAATGAGCAAGCCTATCCATATACCAGTGGGAAAACCGGACGAAG
ATCTGCCAAATGTAATTTTACTATACACAATATTGGCGCAAAGGATTTCGGATATATTGATATTCCTCAAGGAAATGAAG
TTTTGCTTAAAGAGACTGTGGCATTGATTGGTCCAATTAGTGTTGCTATTGATGCTAGTCAATTCAGTTTCAGAAATTAC
CATTCTGGTGTTTATGAAGATCCCAATTGTAGTTCTGTAGAACTTGATCATGGTGTATTGTTAGTAGGCTACGGAATCGA
AAAAGAATCGAACATTCCATATTGGTTAGTTAAAAATAGTTGGAGTACAAATTGGGGAGATTCGGGTTACATAAAAATGA
GACGAAATTCTAATAACATGTGCGGAATCGCTACAGCCGCTTCATTTCCTATAGTGTGAAAGTCTGTCATATTTATTATT
GTTCATTCTCATGATATTCTGTTCATGTTTCAATAGTTGTCCATATCGTACTCTACGTTCTTTTTTATATAATAAAAAGT
TAATAT
>SMED30032925 ID=SMED30032925|Name=Macrophage-expressed gene 1 protein|organism=Schmidtea mediterranea sexual|type=transcript|length=2580bp
AGTCGCGATAGCACGCAACGTCGACATTGCGATGACGTAATCATTGTGAATCTCTACTACAATCGGATCATTCTAATCAC
GTGCATCCTAAAACAATATAAATCTTACAAACGGGAATCCAATTGAACATTTATTTACTCAAGCGTGAAACCAATGATAC
CGTGTAGACTTAATAGTATAATATTGTTTATATTTGTTTATGGAGTTCTATCGGTTTCATGTGAAGGCGAATCAAATAGA
GACCTCCCGTCAGAGAGGCAAGTCCATAAATGTAAACTTACCAACAAGCAATACACATTTGACATTCTTCCGGGGTCTGG
GTGGGATAATCTTGAAAACAAAGAACTCGGTCAAGTTTTAGAAATTTCATACAAGGAATGTTTGGCTTCGAATGATGGCA
GCTTTCTCATTCCAAAAGACGTAATAATAAATCCAATAAAGAAATCTACTGTCGACTTAACATCAGAGATATTTGATCAT
TTCTCAAATGTCACCTCGACAACAGCAATGTCTATAAATGCTGATGCAAGCTTCGCTTCTATAGGCTTTAAAATCAGTGG
ATCATTTTCTTTCGAAAAGAAAAGAATAAAGAACGACCAAATAAAAGAAAAGTCGTCGGTTTCACGAGCTCAATTGAGAC
ATGAATTATACACCGTTTCATTTCAACCTGACTCACAACTCGATCCGGAATTCAGGAACAGAATTCTAGATATTGGAGCG
TTATTGAAAGAAGGAAAAGACACAATTTATCCACAATATTTAGCGGATTTAATTGTCCGTGACTTCGGCACTCACGTCAT
TCGGTCGGTGGATGCGGGAGCGACACTAATAAAGACTGATGTTTTAAAATCGAATTATGTCAAAGGTTTTAAAGGGGATA
CAACTAAGATTGCAGCTGCGGCATCATTCAGTATGACAACAATGCTTTCAGTCAACCTCAAAGGAACCTATGTTGGTGAT
ACCACGAGTCTCAATGAATTCAATAGTAATAGTGCAAGTACAAAAGTCGAAACTTTTGGTGGTATACCGTTTGGTTCGAA
TTTTAAGTTGGAAGATTGGGAGAAAACCCTTGAAACGAATTTGGTAGCAATTGATCGCAAAGGTGAACCGTTGTCCTATG
CTATTACATCAAGAACTCTTCCAGAATTTGACGAGAAAATTATTTACAAAATCGCTACTCTCATCAAAGAAGCAGTCAAT
AGATATTATGCAGCCAACACGAGAGTGGGATGTATGAATATGGAGTCAGAAAATTTCAATGACGACGCAAATGTTGACGG
TGGAAACTGCGAACCTCCTAATACATTTTACAGTTTTGGAGGAACTTTTCAAACCTGCTCAGGCCCTGATTATCAGTGTA
TGGATGTCGCGGCAAAAAACCCAGTGACCAATAGTTATGCTTGTCCTGATAACTATGATACCATTGAATTACAAAGTGGA
TTGAATAAATGTGAGAACGAATGCTCTCATTATGCACTCTATTGGTGTGCCCCTGTTATTCAAAAGATATTTACAGAAGG
CGCCTTATTTGGAGGGTTTTACACGAAAACTTCTCCGAATCCAATTACAAAATCCTATAATTGTCCAGAGAATTATTATG
CATATCAATTTGGAATGACTACAAAGATTTGTTTAGCATTCAATAATGAAAATGCTTTGGAGAATCATTTACCATTTGGA
GGAATTTATAGTTGCAAAGCTGGCAATCCACTTGCAAGTAATCATGAATATGGCATGTTACAAGAATTCGAATATTGGCC
TCAAGGTTGCTCAACCGGATATTCATCACATGTAGCTATTGTCGACAGAGCTTGTCAAATTTCCTATTGCGTTAAAAATA
ACGCCATTAGCATGAGAGAGCAACCGCCTCTCTACCGACCACCTTTTATTGATTTGGGATTTGGACCCAATATGACAGTA
ATTACCTCTCTGGACACTCCAATGGGGGTCCGATTATCTGTTGATGAAAACGGGAAATGGAGTCATAATCACCAATACAA
ATTATCCAATTACAATATTATAATAATATCAGTCATTATGGCGGCAGGAGCCAGCATTGCAATTATTATTATAATTCTTG
GAGTTCAAAGATTCAGCAGAAACCGAAAAGGTCAAGTCTCACCGGAATCTCATGAATTTGATAGCAGATATGATTCTACT
AAAGATGTTGCCAATCTTGCGTTACCCGGCGAGGTTCCACCGTTTCGTGTTATAATTGACGGAGCGAAATGATGCTTGGA
ACCTTTTATTATTTCTGTTGAATCTGATCTGTAAATGTTTTCAGAATATTTTTCTAATAAAATCGTTCTGAAATTTTAAA
GTGATCCTTGATGCTATTAAATGATGGACTTTATTGTTCCCTGAATTGGGTCACCCTTATTCGCATAAATTTCATTATCA
TTTCTTTTTTAGACTATCGATCAATATATTGTCCTATATTAGACATGAATGTTCAGCACTGTCCGTTTAGTTTGTTTAAA
TATAAAACCCAATATCGGCCTTTAGTTCGAAAAAATGTAATAAATAATGAAGAATATAAAAAAAGATTAAATTTATATCA
ACGATTAAATTATTATGTCA
>SMED30007290 ID=SMED30007290|Name=GATA456|organism=Schmidtea mediterranea sexual|type=transcript|length=2033bp
AGGTAAGTGTCGTCATCACAAGTGGCTTTATTATTATCATTTAAATTTTTTATCATATTATCATTGATTGCTTGTTTTCC
TCATTCAAAATTTAACTGATTGTTATCAAGCTCATTTGATAACTCCAAATCTAATTGACACTCATGTAAATATCCCATTC
GAATTTCCGTTCCCGTCCGTAAGATCCACGATCCGACAATGTTTCTCCCATCGGTATTGTCGAATTCTCACCAGAGAAAC
TCACCAATAATGGACATTGTATCACCAAAATCCAACACCAGCAATCTGAATGCGTTGATCACGTATTCGAATTGTCCCAA
CATAGATCTACAGTCGAATGATCTGTCAGTGAACTTGACGAATTCTCAAAACTTCCACGAATTGGATCACAAAACCAATT
ACGGATACAAAGTTTCAAACAACGAGACAAAACTGAATGCTTTTCCATATTTAGCTCAATCGAACTTAACAGATTGCACA
ACAGACATCAGTGGAGATCTGAAAAATCCAAATTCACATCTCAACTATGGAAACGGAATTTCGTTCTCGGAATTTTCGAA
AATGTCCGAAACCAACATCCACGGATATCCGCCATATTGTTTCTCCACTAAACTGTTTCCCATTCCAAATCTACAAGAGA
AATCAATTATCAACGACAACTATTCCGTGAAATCCACCGCCGGAGGCTTTATCACCGAATCGAATGAGCATCAAAACTCT
GTAAATACTGGAAATTCCTACAATTCCAATTTAAGCTCATACAGAACGGTAATTCCCGCTCATTTTCATCTCTTCAATCA
GAATTCACCAGAATCGAACTCGTTTTACAATACGGCTTGGAACAACTATCCGTTTCAAATATCCCCAACTTATCCCATAC
AAAATCTACCGGATTCAACAATCAACTGGACAACAAACAATCAAATTCCAAGCAGCAATTTAGTTTTTGATCCCTGGAGC
TGTAAATCAAATCTTAATCTTCCTCCGATTAATGAATTCGGTTCCAATTGCGATGTCAGCAGAGAGTGTGTGAACTGTGG
AGCTAGCAATACTCAGTTGTGGTCCCGGGATAATTCTGGCTATTATTTGTGTGATGAATGCGACAGGTTCTCTCAAAACA
ACAGTAGAAACCTCGAAAAACTACCAACGAATCAAGTCTCATCAAGTACCGAAAATGAGTTCATGAAAAAATCCAATGCC
AACTTTCAACAATATGGAAAAAGATCCGACCTCGAATGCAGCAATTGTAAAATTACCAAGACTTCTTTATGGAGAAGAAA
CAACGAAGGAGAACCTGTCTGCAATGCTTGCGGCTTGTATTATAAATTACATAAGTCTCTAAGACCTCTTTCAATGAGGA
AAGAAGGAATACAAACAAGGAAAAGAAAACGGAAAACACAAATGAAACCGGTTTCCACTCATATTGCCAATAATCAAAAG
GTGCCGTCTTCAGAATCGTCATCAGAACAAAATCAGTTTATAAACACTGTGAAATCAAGTGAAAATTGCTATAACATGAT
TTCTGAAAATTCTCCAACATATATGCCAAATATATCAGTCCAGCCCTTCAATCACATTCTGAACTCCAGCCAATATCAAT
TCCAATCTACAGAAAATCCCATTAATCAATCATATTTAAATTCCTCAATCAAAAACAGCAATCCCTTTTACGATTCCGAA
AGTATACTTTATTCCTACAACAATTCAAACAAAATCAAAGATGAGCCCCTATTATCCATGGACAACGAACCGAAGCCCTA
TTCCTCAATCAACGAGAAAATTGACGTCAACTCCATAGTGAGAATTAATGAAATCTCAAACAATTCGAATTCGAGTTAAT
TATTCTGATCTACTTTTTAATTATATATTTAAGAACCGTATATACTTCGAAATATAATTTTAATGATAAAATTTTATATC
TAAAAAATGTAATTTTTAAGTGCGAATTATTAAGTATGCATTTAAATAGTGCAATTATTGTGGTAGTTATTTTTATTACT
AAATAAATGTTGTTTGTAAATTCGAGAATTTGT
>SMED30035218 ID=SMED30035218|Name=Gelsolin|organism=Schmidtea mediterranea sexual|type=transcript|length=1573bp
TTATAACAGTCGCTTAATAAAATGATTAAAATCCTTAATTTTTAGTGAATTCAAATAACAAATTTTAAAATCTAATTAAT
AATTAAAATCTGTGACTTTGACAATCGATATGATAAAACAAAAAAAATACAACTGGAAAGACTCCAACTTATCTTTATTT
GGATCTGATACGGAAAAATTAGTCAAAAAAACTTCGTCGGAAACAGAAGCAGCATGGAAATTGGTAAAAACTGTAACTAC
ACCGAGTCTTTTTATTTGGAGAATTGAAAAATTTAAAGTTGTCAATTGGCCTAAAGAAGACTATGGGAAATTTTTCAATG
GAGATTCGTATATCATTTTAAAAATAAAAAAGGAAGAAAATAGTCCTGAATTATTTTATGATATTCATTTTTGGATTGGA
AAAAGAAGTACACAAGACGAGTATGGCACAGCTGCGTACAAAACAGTAGAACTAGATACATTTCTTGATGATAAAGCAAT
ACAGCATCGTGAAGTTCACGGTTTTGAATCTGATTTATTTAAGTCCTATTTTCCGGTTATTACAATAATGAAAGGCGGAT
GTGAAAGTGGTTTCAGACAAGTAACTCCAGAAAAATATGAACCAAGACTTTTACATTTTCACTGTGCTGATCAAAAACAT
GTTGAACTTGTAGAAGTGAAATTTTCAAAAAACTCTTTGGATTCAGGCGATGTATTTATTTTAGACAACGGACTAGAGGC
TTTTCAGTGGAATGGAAAACTATCAAATAAAAATGAGAAAGTTAAAGCAGCACAATTTTTACAGCAATTAGAATCAGAAC
GAAACGGTCGGTGCAAAACAGATGTATTAGACGAAGAAGATACTCCTTTGACGCATAAAATTTACAAATTACTACCTGAT
ACAGCTGTTACTAAGAAGGTTCACAAAGACTTGGCAATTTCCAAACGTCTGTATAGATTATCTGATGAGACAGGCAAAAT
AACGTTTTCTCTAATCTCAGAGGGCAAAACTCCAAAATCTATGCTGAAAGACAATGATGTGTTTATGATAGATAATGGGC
GACACATGTTTGTATATATTGGCGCCGGTGCGTCTCCGGATGAAAAGAAAAATGCAATGTCATATGCACATAATTACTTG
AAAGAAACTGACCATCCATTAATTGGAATTACTTGTCTCCAAGCTGGCCAGCATAGCAAAGAATTTGAGCTTCAATTGGA
CTAGAAAATGATTATGAAATTTTAAATTGCATGGCATTTCAATATTTTTAAATATTTGTGTTTTTACTTAATTTTTACAA
AATGCATGAGTTTATGGTAATTCATTTTCTTAGAAGAGTGAAGAAATATTTTTTTGTTTATGTATTTCATAAATTGCTTA
AATATAATTAAACCTAATGTATATATTGCTGTATTATTTGCTGTTACGAATATTATTGGGCTTGATAGAGCTATCAGAAC
TAACTCGCCATGCTCTTGATCATAACAAGAAATCTACGCAATCAATATATTTGTACCAAAAATCATAACCATTACAATTT
TCACCAACACAAATCAATCTAGTTTTTATTTGTTGCCAGAATCAAATTTTGGG
>SMED30012850 ID=SMED30012850|Name=Spondin-1|organism=Schmidtea mediterranea sexual|type=transcript|length=2699bp
GGTGAATGAAAGATTGAAAGTAAACAAATTTATGAGACATTGACATTATTCATTTAACTTACATCACAGGACGACCAATA
TCACATAATACTTCTTCCTGTTTACAAGGTTGAAGTCGTTTATATAAATCGTCAAAGTCTCAGTTAAGATCATTGATATT
ATGAAAATTTACATCTTAATTTGGAATATCCTTTGGATGTTGCTATGTCTTCATTGGATGATCCAGGAAAGTTTTGCCAT
GAATACTAGAACCAATTGTTTTAAAATGCATAAAAAAACCTACCAAAAAGTCATGATGAAAGATGGAGAATTTCGAATAG
TAATTAGAGACATGAAAGGCAAAATGGCAGATAGATTCGAACCTGGTGAAAGCTATAGAATAATTTTGAATAATGCAATG
TATGGAATGGATTTCAGAGATTTTTCAATATTCGCGGCCTACGAGATGGAGGATTCGTTGCAAATGGAAAGTCATAAAAT
GATTCCCGATGAAGGACTTGGTCATTTTATTATAAATAAAAAATTTGGGTTGGTTGATTATAAATGTCCTAACAGAATGC
ATGGAAATGATAATATGTTTCTAACTTATGTAGAGTCGACATGGATAGCTCCGCATAATCCTATAAATTGTATTCGCTTT
GAGGCTCAAGTTGTAAAATTTGATTTTATGGTGTATGAAAGTATCGGACGTTTAGTAAAAGTTATTTGTCCAACAAAAAT
GTATAAAATGATTAGTGAAGAAAATAAAAACATGAAAATATCTAAACCTTTTGATGAGATGACTCCTATGGTTAAAGAAT
GTTGCGCTTGTGGAGTAGCGTCTTATAAAGTAACATTTATGGGGTTATGGTCAAAGAAAACTCATCCCAAAGATTGGCCA
AAACAAAAGAGCATGGTTCATTGGACCAATCTTATAGGGGCCACACATACTGGCGATTACTTTATATATTATAACGGTGG
TCAAGCAAGCCAGGCAGTTCAAAGTATTTGTTCATATGGAGATAGCACGGTTTTGAAACAGCAGTTCGCAGAAGAGACAA
TAAAACCGCATTTGAAGAGTTATTTTAATACTTCAGGCATGTGGAATGAAGACGAAATTGAACAGAGTAGATCGGGATTC
ATATCGGTTAATCGTACTCATCATTTCTTTAGCTTTCTCACAATGATGGGCCCAAGTCCTGATTGGTGTACTGGAGTAGC
AGCTGTAGACATGTGTATGAATGATTGTACATGGAAGGATGATTTCACCATGGATCTATTCCCATTTGATGCTGGAATTA
AAAACGGTTTCACATATTTCCCAGAAAATTCGGACAGGCAAGACATTCCTGATCCAATTAGAACAATCAACACTACATAT
ATGACCCAATTCCCTTTCACAGCAAATGTTCCAGTTGCCCGGGTAATTTATCAAAAGATCAAACCAAAACACAATTGGGA
GTGCACCAAGATGAATGCAAAACAAGTAAAAGAATTTGATGAAATGAATTCAAACAAAAACGGAAATTCTCAATTCGATG
GAAGTAGTTTGACGAAAAAACGCCGACACATTAACAAGAAAAGTCCCCTTGATGATCCGACTTTGGCCAATATGGCTACA
TTTTTGTGTATAACATCTCCATGGAGTGAGTGGACTAAATGTTCAGTTACGTGTGGCATTGGGACAGAGATGAGATCTCG
AGACATGTTGAAAAATGCTAAAACAGAATTATGTCGTCACTTACCTTTGCTTGAAAGTCGGACATGCGAAGGAAGAAAAC
GTTCGTGTGATTTCTCAGCACCATGTAGCCTTCTTTCCTGGTCAAGATGGGGTCCATGCAATGCAACTTGTGAGATGCAC
GGATACCAACAGAGATCTCGAATGTTTGCAAGATTTGAAGAAAAAGAGAAATGTCTCAAAAACAAAGAAAGACGATTTAA
AATTTTTGAAGAAAAACGGTGCAAAAAAGACACAGCCTTGTGTGACCCAGTTATTATATGTAGTGAGGGCCTAATAATGG
GCAGCAGCTGTGGCAAGCCTGTTATGAAATATTTCTATGATGCTGCCAAACACAGTTGTGTGCCTTTTGAATACCTCGGT
TGTAAAGGAAGTCTGAATAACTTTCCTTCTAAAATGTCTTGCGAAAAGACTTGCTTAGCAGCTGTCAATGCGCTGCCTCA
GTGGAGAAAAGACAAAATGGCTCACTTACAGTTTCAGTCTCAGTCGAATTCAGATAAACGAAAGAAAATGCCAACAATGG
CTAACAAATGTCGGCTTGAAGTAAAATACGGCGGAATGAATTGCGATGGAACAATGGGACCGGACATTCGATATTATTAT
GATAACAGAAATGGACGCTGTAGACAATTTAAATACACTGGCTGTGATGGGAATAGTAACAACTTCCGGTCTTATCACAG
CTGCATTAAGGCTTGCATGCCAGATCAAATCGACTTTATGAAAACTGCTCCGATCAAAGCTTGTCAAGTATCTGAGTGGT
CACATTGGGGTCCATGTTCTACGACCTGCGGTGTGGGAGAGAGATTCAAATGGCGATCTGTGCTTCGGCCGGCATTGAAT
GGCGGTCCAGCTTGTCCAGAGTTATTTCTATCGGAACCTTGTTTCGCGACAACTTGTTGAAAAATTTATATAAATTTTGT
ATCCATTTTTGATGACATTTATAATAATTTTATGGGTAAATATATTTTAGTTCTTCCTC
>SMED30022124 ID=SMED30022124|Name=Gelsolin|organism=Schmidtea mediterranea sexual|type=transcript|length=998bp
TGTACATTAGGATGAATATGGAACAGCAGCGTATAAAACCGTTGAGTTGGACACTTTACTCGACGATAAAGCCATTCAGC
ATAGAGAGGTGCAAGGAAATGAAACTGATCTTTTCAAATCTTATTTCCCAATTATGACAATTATGAAAGGCGGTTGCGAA
AGCGGTTTCCATCATGTTACCCCAGACAAATATGAACCGAGACTTTTACATTTCCGATGCACCGATCACAAGCATGTTGA
ATTGGTGGAGGTGAAACTTTCAAAGCAGGCTTTGGATTCGGAAGATGTTTTAATCTTAGACAATGGCTTAGAGGTTTTCC
AATGGAACGGAAAATTATCCAATAAAAATGAGAAATTTAAAGCCGCACAATTTTTGCAACAGTTGAAATCCGAAAGAAGC
GGACGTGTTAAGACCGATGTTTTGGATGAGGAATTCACTGCTGCCAACCATAGAATATTTAAATTGTTGCCAGATGTTCC
ATTTACAAAGAAAACTCAAAACGATTTAAATATGAGTAAAATGCTTTATCGATTGTCTGATGAAAGTGGAAAAATGACTT
TCTCCTGCATTTCTGAAGGATCAATTCCAAGGTCAATGTTAAAGGATGACGATGTCTACATGATTGACAATGGCAGACAT
TTGTTCGTGTATATTGGAGCACGGGCATCGCAGGAGGAAAAGAAGAACGCGATGTCATATGCCCACAACTACCTGAAGAC
GACTGAGCACCCACTTATCGGAATCACATGCCTCCAATCTGGTCAACATAACAAGGAATATGATCTTCAATTAGAATAGG
ATGTTAAATGATAATTTTGGAATATGTAATAAAAATTAACAAAGCAATTTTTTAAAAACAATAATTTCACAAAATAGGAA
TGAAAATCACAAAAGTATTAAAATTCCATCTATGAATTGAATATCACAATTATAAATTCTTCAACACATAAATCTTGAGC
GAAGCAATAACTTAAAGATTTCAAACATTTACCGCCGC
>SMED30024552 ID=SMED30024552|Name=Myosin heavy chain|organism=Schmidtea mediterranea sexual|type=transcript|length=2149bp
TCACTGGAGTTAATAAACGACAAATTAAGGAAAAATGAACATGAACGATCCAGAAATGCAATACTTGGCCATTGACCGCA
AAGAATTGCTGAAAAGTTTAGATGTTGGGACTTTTGACAGTAAGAAAAATTGCTGGGTCCCCGATGAGAAGGAAATGTTT
GTGGCGGCGGAAATTAAATCCTCCTTAGGAGGTATTTTAAAAGTTCAAACGAAAAATGGAGAAAATCGTGAAGTTAAAGA
AGACGATATTCAACAAATGAATCCACCGAAATTCGAAATGATTGATGATATGGCTAATCTCACTTTTCTAAACGATGCTA
GTGTGTTACATAACCTAAAATCCCGTTATTACCGTGATTTGATATATACTTATTCTGGTCTGTTTTGTGTGGCTATAAAC
CCATATAAAAGAATTCCCATTTACACCCCAAGTATCATGCAGAAATTCAAGGGTAAACGAAAGACTGAAATGCCCCCCCA
TGTGTTCTCGGTATCCGACAACGCATATAATAACATGTTGAGAGATCGGGAAAATCAGTCCATTCTTATTACCGGTGAGA
GTGGTGCTGGTAAAACAGAAAACACTAAAAAAGTCATTTCCTATTTTGCTCAGGTGGCAGCCAATTCAAAAGATGAAGAT
AAAAGCAAAGGATCCCTTGAAGATCAAATTGTACAAGCTAATCCGGTTTTAGAAGCATATGGTAATGCAAAAACAAATAG
AAATAACAATTCCTCAAGATTTGGTAAATTCGTTAGAATTCATTTTGGATAACGGCAAAATTGCCGGAGCTGACATTGAA
TACTATCTTTTGGAAAAATCAAGAGTTGTCTCACAACAAAAAGGAGAAAGAAATTATCATATTTTCTACCAAATATTGTC
TAACAGTGCCAAAAACCTCCATGAAAAATTATTGATAGAACCAAATGCCAGCATTTATTCATTTATTAATCAAGGTGAAC
TAACAATTGACGGGGTGAATGATTCAGATGAATTCGCTTTGACTGATGAAGCGTTCGATGTTTTGGGTTTCACCACTGAA
GAAAAAATGGGACTTTTTAGATGCACAACTGCTATCATGAATATGGGTGAAATGAAATTTAAACAGCGTCCTCGTGAGGA
GCAAGCAGAAGCTGATGGTACTTCAGAAGCTGAAAAAGTTGCATTCTTACTCGGAGTAAATGCTAAAGATTTACTACAAG
CATTTTTGAAGCCAAAAGTAAAAGTTGGTACTGAATATGTTACTAAAGGACAAACTAAAGACCAGGTCTTGTTTGCTCAA
GGAGCGTTAGCTAAATCCATATATAGTCGTATGTTTTCTTGGTTGGTGACCAGAGTTAATAAAACTCTTGATACGAAAGT
GAAAAGGCAATGCTTTATTGGTGTGTTGGATATTGCCGGTTTTGAAATCTTTGAATATAACGGATTTGAACAACTGTGCA
TTAACTACACAAATGAAAGACTCCAACAATTTTTCAATCATCATATGTTCGTATTGGAACAAGAAGAGTATAAGAAAGAA
GCGATTGACTGGGAGTTTATAGACTTCGGATTAGATTTACAAGCTTGCATTGAACTTATTGAAAGACCAATGGGTATTCT
GTCAATTCTTGAGGAAGAGTGTATGTTTCCTAAAGCAACCGATGAGACTTTTGTTGCAAAATTGTATGAAAATCATCTGG
GTAAAAGTCCTAACTTTGGAAAACCAAAACCAAATGGTCAAGCAACTGCCCATTTTGAGTTACATCATTACGCTGGCTCT
GTTGGATATTCAATTACTGGATGGTTGATAAAAAATAAAGATCCAGTCAATGACAGTGTTCTTCAAACTTTAGGAGCTAG
TAAAGAAGCTCTTGTTGCTCAACTTTTTGCTCCTGCGGATGATGATGCTACTGGAAAGAAAAGGAAAGGAGGCAGCTTCT
TAACAGTATCGTTTGTTCACAGAGAATCTTTGAATAAACTTATGAAGAATTTACAAAGCACTAGTCCACATTTCATTAGA
TGTATTATTCCCAATGAATTAAAACAATCTGGATTCATTGACTCCCACTTAGTCCTACATCAACTTCATTGTAATGGTGT
GTTAGAAGGTATAAGAATATGTAGAAAAGGATTTCCAAATAGACTTATTTATTCTGAATTTAAACAAAG
>SMED30032088 ID=SMED30032088|Name=Spondin-1|organism=Schmidtea mediterranea sexual|type=transcript|length=1776bp
TTACCTGAAAATAGATTAAATGTTAAGAGTCTCTTTACGTCAGGCTGGTTAAATGGGGAAGAACAGCTAAATGAAGAGCG
TTCTGGATTCGTTTCTGTTAATAGAACTCATAATTTATTTAGTATGATTTCAATGATGGGACCAAGTCCGGATTGGTGTA
CCGGAGTGTCAGGAATCGATCTTTGTATGGACGGATGCAAATGGAAATCAAATCTAACAATTCCTCTTTATCCTATCGAC
GCTGGAATACAAACTGGAGAGACCTACAAACCAAATAATGTCACAAGACTTGATGAACCCGACACTATTAGAGAAATTAA
TAAAATTTGGATGAAAAGTAATCCTTTTACTCCAGATACTCCAGTAGCAGAAATCACCCTCACCAGAGTAGAGCCACAGA
AAGACACACAATGTGTTAAAACTCCAGTTAAGGGTGTAAATGAATTTCAAAATGCACAAAAAGAGTCTAGATCTAACACG
CTCGGTAGAAATGGTGCCGTGGAAAAATATAATGTTCTAGATGATCCTCAGTTAGCTCAAATGGCAAAAGCCTTTTGTGG
ATTTTCCGAATGGACCAGCTGGTCACCATGTTCCGTAAAATGTGGAGAAGGTGAAACGTCTCGTACTAGAAATTTGATAA
AAAACTTGAAAAATGAATTGTGTATTCAATTGCCAAGAAAGGAGCAAAAAAATTGTGCCGGTGCTCGATTGGAATGTGAT
CTCTCAAAGAAATGTAGTCTATTACCTTGGAGCTCATGGAGTCCGTGTTCTGCATCTTGTACAACTGAAGGGAAAATGAT
CAGAATTAGAAAATTTGCGAGGTCAAAAGAAAAGGACGAATGCATGAAAAACGTCAAACCAGAAATTATTCTTCAACAAG
TGAAAACTTGTAAATTGGATAAAACGGACTTCCGATGTGATCCGATATCAATTTGCAGCGCCGGTAAATTGGCACCTTGC
ATCAATGGTAAACCCAAGACGAGATTTTTCTACCACATCAAAACTGGAAAATGCGAACCGTTTAAATACTATGATTGCAA
AAAAGGAAATCTGAACAATTTCCCAACTATAGATTCATGTGAAAATGTTTGCTCTAAAGTTATTCAGGAATTACCAAAAA
TAAGACGATCTAATTTAGCCTTGCTTCAAGCAAGAACATCTTATTCAGCTGAAGAGAGTCTTTCGCAAAAAGCCAAAAAC
ATTCCGGAATTCTGCTTTCATGAAGTGGATTCCAGTAAGACCTGCATGAAATCCAAACCAAAGACAATCTGGTATTATAG
TCATCGAAATCGAAAATGCCAATCACTGGTGGATTATGATTGTAAAGGAACTTTAAATAAGTTTTCGACTTTCGACGATT
GCATGAAAACCTGCCGACCTGATTCACAACTCGAATCATTGAAACTTGATAAAGTAGATTGTCAGGTCTCTGAATGGAGT
TTATGGAGGCCATGTTCAAAGTCATGCGGTTGGGGAACTCAATACAAGAAAAGAGTCATTCAAAAGGATTCATTGAATGG
GGGCTTACCATGTCCGGTGCTATATGAATCTCGAAAATGCTTTTCCAAACCTTGTTAATCTGTCAATATTTATTTAAAAA
TTGTTTTTTGTGAATTATAATTTCTATTTCTTAATCTATTTGAAAATGTAATAAATATTACTTTGCGTATTTGATTTAGT
TTAAAGTTTTCTCATTGGTCAATCCGCCGTTGCTAACCAATGAGATCGGCTACTACATCAATTGCGATAATTGACAGTGG
TGAGCAAACCTTTAAG
>SMED30022389 ID=SMED30022389|Name=Proliferating cell nuclear antigen|organism=Schmidtea mediterranea sexual|type=transcript|length=915bp
GTTATTAACGTTATTGGACTATAGTTTTATTATCATAAATTTTTGAATAAAATTGAGATCTTCTTTTAATAATGCTATTT
GAAGCAAAATTAAATAAAGTAGATGCATTTAAAAAGCTGATTGATTCGGTTAAAGAATTAATCCAAGAAGCAGTTTTTGA
ATGTTCATCCAAAGGTATTGAATTACAAGCAATGGATAGTTCACATGTTAGTCTAATATTTCTTTTAATTAAAAAGCAAT
CTTTTTCGTCTTATAAATTTACGAGAAATGTATCTTTGGGTGTAAGTCTTGTAAGTCTATGTAAAATACTCAAACTTTCT
CAAATTAATGATTCAATTACAATTAAAGTTGATAATGAATGTGAAACAGTCGAATTTGTTTTCGAATCATCAGATAGTGA
TCGAATGTCTGAATTCAGTTTGAAGTTAATTGACTTGGATATTGACTGTTTAGCTATTCCGGAATCAGACTATGATTGCG
TAGCAACAATCCCGTCTGTAGAATTTAACAAAATATGCCAGCAATTAGCTGTTATTGGAAGCACTGTAACAATTGTAGTA
ACTAAACAGTCAATTCAATTTTCTTGCTCCGGAGACATGGGGAAAGGGAAAATATTATTAAATCAAACTGTAAATAAACC
GAGAATAGAAGATTCTATTAATATTGAAGTTGTTAATCCATTATCAATGGTTTACTCGCTTCGATTTTTTCATATATTCT
CGAAGGCCTATTCGCTTAGCAACAGAGTTTCTTTATCTTTTGCTAATAATTTCCCAGCTTTGGTGGAGTACAGCATGAAT
GAATTGGAAGGTCATCTACGATTTTACTTAGCTCCAAAATTAGATGATGATTGATTTTGCTCTTTTTCTAGTTTTGGGGT
GATATTTTAATTGTAGTGCTTTTTTAGTATGGTTA
>SMED30012543 ID=SMED30012543|Name=Spondin-1|organism=Schmidtea mediterranea sexual|type=transcript|length=2035bp
CTCCAATGAAATGTTTACTTTTATAAATGACAAGCCTATTGTAAACAATGGAATGAAGACGGCTTACAAACAATGTTGCG
CCTGTGGAACTGCAAGTTATCGGGTGACTTTTCATGGGTTATGGAACAAAACAACTCATCCAAAAGAATGGCCAGAAGAA
AAAGGTCAGCTTCATTGGACAGATCCTATTGGAGTCACCCATAATCATAATTACTTTATTTATCATTTGGGAGGACAAGC
TTCATTTGCGGTTGGTAGTGTATGTACAACAGGAAATTCGAGATCTTTGAAAGAGGAATTCACCTTACCTGAAAATAGAT
TAAATGTTAAGAGTCTCTTTACGTCAGGCTGGTTAAATGGGGAAGAACAGCTAAATGAAGAGCGTTCAGGATTCATTTCT
GTTAATAGAACTCATAATTTATTTAGTATGATTTCAATGATGGGACCAAGTCCGGATTGGTGTACCGGAGTGTCAGGAAT
CGATCTTTGTATGGAAGGATGCAAATGGAAATCAAATCTCACTATACCTTTGTTTCCGTTTGATGCTGGAATACAGACTG
GGGAAACCTATAAACCTGAATCTATTACTCGGCTTGATGAACCGGAGCCTATTAAAGAAATCAGTAAAATTTGGATGAAA
AGTAGTCCGTTTACACCGAATGTTCCTGTTGCTGAATTAATTCTGACAAGAATCTCTCCAAAGACCGACGAAAAATGTTC
TAAATCCCCTATGAAAAATGTTGATGAATTTAATAAGTCAAATGAATTAATAAAACAAAATTATCGCGGTAAAAATAAAG
GAATAGCAAAATATAATGTTCTTGATGACCCCCAGCTGGCTCAGATGGCAAAAGCTTTTTGTGGATTTTCTCAATGGGCT
AGCTGGTCTCGGTGTTCAGTGGATTGTGGAGTTGGGGAATCTTCTCGTAAGCGAGAGTTGATCAAGAATTTTAAAAATGA
ATTATGTGTCCAGCTGCCAAGGAAAGAAACCAGATATTGCTATGGTCGACGAAGAAATTGTGATTTTTCTATGAAATGTA
GTCTTCTTCCGTGGACTGAATGGACTAAATGTACTGCTACTTGTAAAGAAGCAGGAAAGATAGCAAGATCGAGAAGATTC
GTGAGGGAGGCGGAACGAGAAGAATGTTTGAGAGAAGTGACACCAACTACTAGACTATATGAGGAAAAGCCATGTAAACT
CGATCAAACTGACAGCCGCTGTGATCCAGTGTCAATTTGCAGTGCTGGAAAATTGCCTCCTTGTACTAGAGGTAAACGTC
AAACAAGATATTTCTATCACGTCAAGACTGGAAAATGTGAACCGTTTTTTTATTTTAATTTTTGCCAGATTGGCCTAAAC
AACTTTCGCACTGAGGTAGACTGTGAGAAAATGTGCGTTAAAGCCATTGGTCAGCTGCCTGGAATTAAAAGATCAAATTT
AGCTCTTTTGCAGTTTCGAACTTCTTATTCAGGAGACCTTTCTCAAAAAGAAACTAAAATACCAGCTCAATGTTTCAACG
AAATAGAATCAGTAAAGACATGTATTCATAAATCTCCAAGAACCGTATGGTACTACAGTCACCGAAACCAGAAATGTCGT
TCTTTGGTGGATTTCGGTTGTCAGGGAAGTTTGAATAAGTTTTCTTCTTTTGAACAATGCATGAAGATTTGTCGACCTAA
TTTACAGAGTCGTTCTTCAACATCTGATAAAGTAGACTGCCAGGTCTCTAATTGGAGTTCCTGGAGGCCTTGCTCTAAAA
GCTGTGGCAAAGGAACACAGATCAGAAAGCGAATAATTTATAAAGATTCTCTTAATGGTGGAGAACCTTGTCCACTTCTT
TACGATTCTAGAGATTGTTACGGTATATCTTGCAATTAGAGATAGCCATAACTATTTTAGTTTATAAAAGCATTCATCCA
GGCCACAGTTGAAAAGCATTTTAATTATTAATAAATTTTAGAATTGTTAATTTTAGTATTTTAGTATATTTTTGCTGTAT
TTAGTGCAAAAAAAAAAGACGGGTATAAAAAGGTG
>SMED30003202 ID=SMED30003202|Name=Eukaryotic initiation factor 4A-like Protein|organism=Schmidtea mediterranea sexual|type=transcript|length=1324bp
ATTAATAACAATGGAAAATCCAGGAGAAATAGAAAGTAATTACGACAAAATAGTTGAATCATTTGAAGATTTAAACTTAA
AAGAAGATTTGTTAAGAGGAATCTTTGCTTATGGTTTTGAAAAACCTTCTGCTATTCAGAAGCGTGCTATTTTGCCAGCT
ATTGAAGGTCACGATGTCATTGCCCAAGCACAATCTGGAACCGGAAAAACTGCCACTTTCAGTATTGCTGTTTTGCAGAA
TATTGACACTTCTATCAATTCTACACAAGCTATTATTCTAGCACCTACCAGAGAACTTGCAGTTCAAATCCACAAAGTTA
TAATGACCCTTGGTGACTATATGGGAGCTAAATGTTGCTCATGCATTGGTGGAAGAAATGTCCGTGACGACATTAACAAT
TTGGAAAGTGGAATGCATGTTGTTGTCGGTACTCCAGGACGAGTTAAAGATATGATAAAGAAGGGCCATATTAAATCAGA
TTTCGTGAAAATGTTTGTCCTTGATGAAGCCGATGAAATGTTGAGTCGAGGTTTTCAAAATGACGTTTGTGAATGCGTTC
AATATTTATCTGCCGAAACCCAAAATTTCATTCTTTCTGCAACAATGCCAGATGAAGTTTTAAAAGTCAGTAACGAATTC
ATGAATAATCCAGTGAAAATTCTCGTTAAAAAAGAGGAATTGACTCTGGAAGGTATTCGTCAATTCTACGTTAACGTTAG
CCAAGAAGAATACAAATTTGATACCTTATGTGATTTATATGAATCTCTGACCATCACCCAAGCTGTCATTTTCTGTAACT
TCAAAAACAAAGTTATATTTTTAAATGATAACTTGAAAAGTAAAGGATTTGTTGTTTCATCAGTCCACGGTGACATGGAG
CAACAAGCTAGAGACCAGATTATGAAAGAGTTCCGAAGTGGATCAAGTAGAATTCTCATCACCACAGATTTGCTATGCCG
AGGTATTGACGTCCAACAAGTCTCAATGGTCATTAATTATGATTTGCCTAAAAACAAAGAAAATTATATTCATCGAATTG
GCAGAGGAGGCCGATTCGGAAGGAAAGGAGTTGCACTAAATTTTGTCACAGACAAAGATATTGCTGAAATGCAGGAGCTG
GAAAATTTTTATAACACCAAGATTGAAGAGATGCCCATTAATTTATCTGAATTCTTATAAATGATTTACCGCTATTTTTG
TAAATAAATTTTAATTTACAATTAAATTAGGTTATTCATAACTTCCTTTGTTACGTAATTTTTGCTATCTTATCTATTTT
TGTTAGTAATGTATTTATGCTGCGTCACATTTTTGTTGCAAATT
>SMED30019745 ID=SMED30019745|Name=Macrophage-expressed gene 1 protein|organism=Schmidtea mediterranea sexual|type=transcript|length=2146bp
CAAAGTTTAAAAATTTATTGTAAATGTATATTCAAATATTACTAATAATTAGTCTTTTCAAAATATTAGTGAAAGGATCA
AATGAAGTCAATACTATCAAGGATGCGTATCAATTGTGTCAGTTGATGCCCAATCAAAGAACGTTCGATGTTTTACCTGG
ATCAGGATGGGATAACCTGGCCAATATCGACATGGGAAGGATACTTGATTTAAACTTTTCAAAGTGTTCATTTTCGAATG
ACAACAAATATCTGATCCCGAATCAACTGGCCATTTATCCAGAAAAATCTTCCCAAGTTGAAATCTCAAGTGAAATATTC
CGTCATTTCTCAAATTACACATCAATGTCTTCGCAATCTATCAATGTTGGTGCTGAAGGTGGCTACGGAATATTTAAAAT
TGGGGGATCTTTTTCCACGGAAAATACAAAGATCAAAAGTGATCAGATGGAAACGAAATCCATTTTCACCAGAGTTCAGT
TGCGATATTTGGGGTTCCAGTCAATGTTTGAGCCGGATACGCCTCTCTATCCGACATTCAAGAACAGAATCATGGACATT
GCTGGATTAGTTGTTGAAAATAAAGACTCACTTTATCCTGAGTATTTGGCTGACATGATTGTTCGAGACTACGGGACCCA
TGTAATCCGTGCTATAGATGCTGGGGCGATTTTGTCAAAAACCGATGCTCTACAATCTAAATATGCAAAATCTTACCAAT
CTGACAGCACTACAATTTCGGTTTCTGCGACTGCGTCCTTTCTAAATCTATTCAGTATGAGTTCTAAGTTTCAATCAACA
ACTAGCCAAGAGAAAGTCAATGAATATAATGAAAATATATCGAAAACTCACATTGATACGATTGGAGGGGGATTATTTGG
AGCTAATACGACTTTAGCAGATTGGGAAGAAACAGTCATGAATAATTTAGTTCCAATAGATCGTAGGGGTGAACCCTTAT
ACTATGTCATTACTCCCCGAGCGTTACCGGAACTTGATGACGAAACCGTAAATCAAGTTTATCAACTAGTGAAGAAGGCA
ATTGAACGATATTATTTTAGAAATGGCCGCAAAGGTTGTCTCGATATCATGTCTCCTAATTTCAATCCAGATGCCAATAT
AGATTCGGGTAATTGTGAACATCCTTCAGTAAATTTTACATTTGGAGGAGTTTATCAATCATGTGAGGGACCCTCTTGTA
GTTCTAGTGTTCAGGAAAATCCTTTAACTGGAACCTACTCGTGTCCTTCTGGATTTAGTTCGTTAGAACTTCATCGTGGG
CTGGTAAAGTGTATACAAATATGTCATGGGTGGTGGATCTTTCGTAACTGTGAAACAAAATGCTCATATTATACACTTTA
TTGGTGTCATCCAAATGAAAGTGGATCAAATTATAATGGGTATCAATTTGGAGGCATTCGGCAAGGAAGTTGTCCCATTG
GATTTATTCCACTTAAGTTTGGATTAAATATAGAAATTTGTGTGACCATTGATACAGATCCAAACAACCCATTTGTAATC
CAATTTGGTGGTTTATTTAGCTGTAGTGCCGGTAATCCTTTAGCCAAAGAATATGTAAATGGAAAAGCAAAATCATTTCA
AAATTTAAAGATGAATGACATAAAATATTGGCCTAAAACATGTGCTAAAGGATTTGTGGCTCACATTGCAACCATTGAAC
AAGGATGTCAGATTTCCTACTGTACCGAAACCAATAAAGTTTCTTATGGTGCTTTTCCTCGCTTAATCAAGCCACCATTT
ATTTCAATAAATTACAATCCAAACAGTACAAAAGTGTTAGTTTTTTCCAATTTAAAAGGAGAAACTTTGATCAAAGATAA
CTTTGGAACATGGAAAGTGGATGAATTGAAAAAGCAAAATAGAGATTTGCTAATAGGAACAATAGTTGGCAGTTTGTTAT
TAGCAGCTTGTATTTGTTTCGTTATCTTTTACATTCAAAGAAGATATTACATTAATAGATACAAGCTGATTAATAGAAAG
AAGTTCACTGATGAAGAAAGCATGACAAATTATGGCTCAATCAACGGAACGGAAATAGCGTAGCCAATCAGATAATTACA
AAAGCCTATGTGAACAATCCTTTTATATTTATCTTTATATTTTATCTCACTTTTCGATCCAGATCG
>SMED30014940 ID=SMED30014940|Name=Gelsolin|organism=Schmidtea mediterranea sexual|type=transcript|length=530bp
CAAATAAATTTTCGTTTCGTAATTATTTAAATTTCACATGTTTTTTATTTAACTATAAATTCTGGAAAATAAATTAATTT
TACATTCAATTAATTCAAACAATGAAATTAGATTTTAAAAGTAAATCATTTCTAGAATTTTTTGTTTTTTAAACTTTCTA
AAAAGGAAACAACTACGGAGCAGTCGAAGAAGTTCTTGTCACCGGACACAGAAAGTCTGACAAAACCGGAATGATAAAAC
AAAAAAACTACGATTGGAAAGATTCCAATTTGGCTTTGTTTGGATCAGACACGGAAAAATTAGTAAAAAAAACTTCCGCT
GAAACCGAACCAGCATGGAGAGTTGCTAAGGCAACTACGGTTCCAGGTTTATTTGTTTGGAGAATAGAAAAATTCAAAGT
CATCAGTTTACCAAAGGAAGAATTCGGAAAATTTTACAATGGAGATTCTTACATTATTTTGAAAATCAATAAAGAAGAAG
ACAGTCCGGAGTTGCTATACGATGTGCATTTTTGGATTGGAAAATATTCT
>SMED30011876 ID=SMED30011876|Name=Macrophage-expressed gene 1 protein|organism=Schmidtea mediterranea sexual|type=transcript|length=1880bp
AACGGAAATACAAATAAATTCTCGATTTGTCAACTCTCTAAAGTTCAAGAGTGCTACAAAGAAAAAAATAGATCACAAAT
GTCCTCTGAGCAATAATCAAACTCAATTCAACACTTTTCCCGGGTTGGGGTGGGACAATTTGGTAAATCAAGAAATGAAG
CCCGTTCTGAAGATCTCATTTTCCAAGTGCGAAGAGTCCAGTGATGGAGTTTATCTCATTCCAGACATGACGACCATTTT
TCCATTAAAAAAATCAAAATTTCATATGAATTCTGATTTATTCTCCAGCTCTTCTGAATATGTTTCTCTCACTTCTCAGT
CCATTAATGAAGATATTAATTTCTCTGATTTCCCTACTTTAATAAATGGCGCATTTTCAAACGAAAAGACCTCTTTGAAA
AGGAAAAAAATAAAGAAAAATTCTATTTTATCTCAAGTTCAATTGAGATACGAAGTTTATAATATTTTGTCTCAACCGGA
CAGTGATCTCTCGAGGGAATTCAAAGACCGAATTGTTGAATTAACGAAATACTTGGACGACGAAAAGAATTTGTTATATT
TGGATTACTTGTCGGATTTGATCATAAGAGATTTCGGAACGCATGTCATCAAGTCCGTGACAGCCGGGGCAATTCTTGCC
AAAATTGACGTTCTCAAACCGGAATATGCGTCAAAATATAATAAAAATCAAGGGGAAATTAAATTAGCAGCCGCCTCAAG
CATGGCTGAAAACGTCGGTACGAATCATGAAAAAGATGAAAGATCAAAAAAATTATTAAAAGAATATAAAGACAATTGTA
TCAGTTCGACAATGAAAGCAATCGGTGGAATGCCAATTGAGTCAAATTTCAATTTGCAAAAATGGACAAATACCATAAAG
TCGAACTTAGCACCCATCGACCGACAAGGTGACCCACTCAATTATGTGGTGACAAAGAAAAGTTTACCGGAATTGAAAGA
TAAAGCAATTTTCGATATAAATAAATTAATTAGTAAAGCAATAAACCGTTACTACAGAGCAAATATGGGATCAGATTGTA
CGAATATTGGTTCTATGAGTAATGAAAATAATGGAAATATTAATAATAAAATGTGCGAGCAGTCAACAGAAATTAAAAGA
TTTGGTGGAATATACCAAACATGCGATGGTGAATCGTGTAGTGAATACAAACAGGATAATCCTGCGACTGGAAATTTAAG
CTGTCCAAAAGGCTACAATTCGTCTATCCTTTATAAAACCATGAACATTAGTAAATGTAAAGAAAATTGTCGAAGCTTAA
CTTTGTACATGTGTTATGCAAACAGCAATGGATTCAATGATCTTGGTTTTTCTTTTGGTAGAATAATACCCAGATTCAAG
CCGAAGTCCGATTGCCCTAAATATCACATCTCGCTACGCTTTGGTGCGCAAAACATTTTATGTGTCACTTCAGGTAGAGA
GCAAGTAGAAGAAAACAGCTTACAATTCGGTGGTTTTTACAGCTGTTTGATAGGAAATCCGTTTACTATAAAAAAACAAT
CAAACGACAGTGCGAATGGCATTTATGAAAAATCATGCCCCCCAGGATTTACAAGTCAACTTAGTCAAGTCGATTCGGAT
TGCCCGGTTTATTATTGTACGAGAATATACAAAACCATCAGCAGAGTCGTTTTATTAAGACCGCCATTTATCAAAATTGA
TTGGAATTTAACTGCTACTAGCGCTGAAACATTCGACAAAATTGACGGTAAAACTGAGATGAAAAAGATCTGGAATAATA
AAGCGTTTCGGTATTGTGTAATTGCATTAGCAATTTTCTTATTTGTTTTCTTACTTCTATTTTTAATATTGTTCCATCTG
AAATTTTGCAAAAAATAAATGTTAACTAATTGGTGAGGCC
>SMED30016281 ID=SMED30016281|Name=Myosin heavy chain|organism=Schmidtea mediterranea sexual|type=transcript|length=5986bp
CTAAAATTTAGTTGATTTCGATTGTTTGAATAAAAATATCTAATGGACCCATCAGATCCTGATTTTATCTACTTGGGAGT
CGACAGGAAAGCTTTATTGAAGCAATATGGCGACTCGTTTGATAGTAAAAAGAACATGTGGATACCGGATGAAAAGGAGG
GGTACATCGCAATTACCGTTGAAGAAACAAATGGCGATAAAGTAACTGTTAAAACCGATAAAGGAGAAACTAAAGTCGTC
AAGCAAGACTCCTTGGAACAAATGAATCCCCCCAAATTCATGATGATTGAAGACATGGCGAATTTAACATTTCTCAATGA
TGCCAGTGTATTTGATAATCTGAGACAAAGATATTACAAAAATCTGATTTATACTTACTCCGGACTGTTCTGTATTGCAG
TGAATCCTTATCGAAGATTTCCGATTTATACGTCCCAAGTAATTATGAAATATAAAGGAAAACGTAGAGGAGAAATGCCA
CCGCATATTTTCTCAATTTCGGATAATGCTTACTCAAATATGCTTGTCGATCGAGAAAATCAGTCTGTTTTGATTACTGG
AGAAAGTGGTGCTGGGAAAACTGAAAACACGAAAAAAGTTATTACTTACTTCGCCAATGTAGCAGCAGCAAGTAAAAAAG
ATGATGACGATCCATCGAAGAAAAAAGGTTCATTGGAAGATCAAATTGTACAAGCCAATCCTGTACTGGAGGCATACGGA
AACGCCAAAACAACTAGAAATAATAATTCGTCAAGATTTGGAAAATTCATCCGAATTCATTTTGGCACTAGTGGAAAAAT
TGCAGGAGCTGATATTGAGTTTTATTTGTTGGAGAAATCTCGTGTTATCTCACAGCAAAAAGGAGAAAGAAATTACCATA
TTTTCTATCAATTGCTTTCGGCAGGAGGGAAAAGTTACCACGATAAACTTTTGGTTACTGCTGATCCTGCTCTGTTTTCT
TTTATCAACCAAGGAGAATTGACGATTGACGGTGTTGATGATGAGGAAGAAATGAAACTTACTGATGAAGCCTTTAGTAT
TCTTGGGTTTTCCGATGAAGAGAAAATGAGTTTGTTTAAATGCACTACTTCAATCATGAATATGGGAGAAATGAAATTTA
AACAAAGACCCCGTGAAGAACAAGCCGAGGCTGATGGTACAACAGAAGCTGAAAAAGTCTCTTTCTTACTTGGAGTGAAT
GCCAAAGATCTAATGCAAGCAATTCTTAAACCCAAAGTCAAAGTAGGAAACGAATATGTAACGAAAGGTCAAAGCAAAGA
TCAGGTTTTATATTCTCTTGGAGCCTTAGCAAAATCTTTATATAACAGAATGTTTGCTTGGTTGGTTACTCGCGTGAACA
AAACGCTGGACACAAAAGTCAAAAGACAGTTTTTCATTGGTGTCTTAGATATTGCCGGATTTGAAATATTTGATTTCAAT
GGTTTCGAACAAATTTGTATCAACTATACTAATGAGAGACTCCAACAATTTTTCAATCACCATATGTTTGTTCTTGAACA
AGAGGAATATAAAAAAGAGGCCATTGACTGGGAGTTCATAGATTTTGGAATGGATCTCCAAGCTTGCATTGAGTTGATTG
AAAAGCCTATGGGTATTTTGTCGATTTTAGAGGAAGAATGTATGTTTCCCAAAGCAAGCGATCAAACATTGAAAGCTAAA
TTGTACGATAATCACTTGGGTAAAAGTCCGAATTTCGGAAAACCAAAACCACCAAAACCAGGACAACACGAAGCCCATTT
CGAATTACATCATTATGCTGGCAGTGTTCCATATAATATTACTGGGTGGTTGGAGAAAAATAAGGATCCATTGAACGAAA
GTGTTGTTGGATTACTCGGAGCATCTAAAGAATGTCTAGTAGCTTCTTTATTTGCACCGGCTGAAGATACAGGAAAACCC
GGAACCAAGAAAAAGGGTGGATCCTTTATGACTGTGTCATATATGCACAGGGAATCATTGAATAAATTGATGAAAAATCT
AATGAGTACAAGTCCACATTTTATTCGTTGCATTGTCCCCAATGAATTCAAACAACCAGGAGTTGTTGATGCGCATTTGG
TTATGCACCAACTTCATTGCAACGGTGTATTGGAAGGTATTAGGATTTGCAGAAAAGGATTCCCAAATAGAATGATCTAT
TCTGAATTCAAACAAAGATATTCAATTTTGGCACCGAATGCCATTCCACAAGGATTTGTTGAAGGCAAACAGGTCACTGA
TAAAATCCTCACAGCCATTCAACTGGACACAAATCTTTATCGCTTAGGAAATACCAAAGTATTTTTTAAAGCTGGAACAT
TGGCTCAACTTGAAGATATAAGAGACGAGAAACTTTCGTCGTTGATATCGTTGTTTCAAGCTCAAATTAGGGGATATTTA
ATGAGACGGCAATACAAAAAACTTCAAGATCAAAGAGTTGCTCTGTCTTTGATTCAAAGAAATATTAGAAAATATTTGGT
TTTGAGAACATGGACCTGGTGGAAATTGTTCACTAAAGTAAAGCCTATGCTTAATATTGCTCGCCAAGAAGAAGAAATGA
AAAAGGCAGCCGAAGAATTAGAAAAGCTGAAAGAGGAATTTGAAAAAACTGAAAAATATAAGAAAGAACTTGAAGAACAA
AATGTTACTTTACTTCAAGCAAAGAACGATTTATTCTTGCAGTTGCAAACTGAACAAGATAGTTTAGCTGACGCTGAAGA
GAAAGTATCTAAATTAGTTCTACAAAAGGCTGACATGGAAGGAAGAATTAAAGAACTTGAGGACTCACTTTCAGAAGAAG
AAAATTCAGCTTCAACATTAGAAGAAGCAAAAAAGAAACTCAATGCTGACATCGATGAATTGAAGAAAGATGTTGAAGAT
TTGGAATCATCTTTACAAAAAGCAGAACAAGAAAAAGTTGCAAAGGATCAGCAGATTAAATCCCTCAACGACAATGTTCG
TGAAAAAGAAGAACAATTAGCCAAAATGCAAAAGGAAAAGAAAGCTGCTGATGAATTACAAAAAAAAACAGAAGAATCAT
TGAAAGCCGAAGAAGAGAAAGTCAGTAACCTCAATAAAGCTAAAGCTAAATTAGAACAAGCCCTGGATGAAATGGAAGAA
AATCTTGGTCGAGAACAAAAAATTAGAGCAGATGTTGAAAAAGCAAAACGAAAAGTTGAAGGTGAACTTAAACAGAATCA
AGAGTTATTAAATGACTTGGAGCGCATAAAATCCGAACTTGAGGAACAATTAAAACGAAAAGAAATTGAATTGAATGGTG
CTAACTCTAAAATCGAAGATGAATCTAATTTAGTTGCAACACTTCAACGGAAGATCAAAGAACTTCAAGCCAGAATCCAA
GAATTAGAAGAAGACCTTGAGGCGGAAAGACAAGCAAGGGCAAAAGCTGAGAAAGCTAAACATCAATTGGAAGCGGAACT
TGAGGAAATCAGTGAAAGACTAGAAGAACAAGGAGGTGCAACTCAAGCCCAAACAGATTTAAATAAAAAACGAGAAGCTG
AACTCATAAAATTAAAGCGAGACTTGGAAGAGGCTAATATGCAACACGAACAAGCCTTGGTGCAAATGAGAAAGAAACAA
CAAGATACTAGCAATGAATTCGCAGATCAGTTAGATCAATTGCAAAAATCAAAATCAAAGATTGAAAGAGAAAAGAGTGA
ACTTAGAGCAGACATTGAAGATTTATCAAGTCAGCTGGAATCACTGAATAAGACCAAGGGAAATCTTGAAAAGTCAAATA
AAGCATTAGAAACCACAGTTTCAGATCTTCAAAACAAACTCGATGAATTAAGCAGACAATTGACAGAAGCAGGAAACGTC
AACAATCGAAATCAACATGAAAATAGTGAAATGCACAAAACCTTAGAAGACGCCGAATCACAAATAAACCAACTCGGAAA
GGCTAAACAACAACTCCAAGCGCAGCTAGAAGAGGCTAAACAAAATTTGGAAGATGAATCACGCGCAAAAGCAAAACTTA
ATGGAGATTTAAGAAATGCGATGAGTGATTTAGATGCATTACGGGAATCATTGGAAGAAGAACAAGAAGGCAAAAGCGAT
GTTCAAAGACAGCTTGTGAAAGTCCAAAATGAATTACAGCAACTTAAAGCAAATTCTCAAGGTACTGGAGGAGTGAGAAC
AGAAGAAATGGAAGAATTCAGACGAAAGATGAATGCCAGAATCCAAGAACTAGAAGAAGAATCGGAATCAAATAAATCTA
AATGCAGTCAATTAGAAAAAATGAAATCCAGACTTCAAGGAGAAATTGAAGATTTAGTGATAGATGTTGAAAGAGCCAAT
GGATTAGCATCGCAACTGGAAAAGAAACAGAAAAATGTTGACAAATTAATAGGCGAATGGCAAAAAAAATATTCAGAAAG
TCAACAGGAACTAGAAGTATCACTGAGAGAATCAAGAACAGTTTCCGCTGAAGTTTTCAAATTGAAATCACAATTAGAAA
ATTCTCAAGATCAAATCGAAAGTTTAAGAAGAGAAAATAAAAATCTATCTGATGAAATTCATGACTTAACTGAACAATTG
GGAGAAGGAGGACGAAATGTACATGAAATTGAAAAGACTCGAAAAAGAATTGAAATTGAAAGAGATGAATTACAACATGC
ATTAGAAGAAGCTGAATCAGCTTTGGAACAAGAAGAAGCTAAAAGCCAAAGAGCTCAGTTAGAAATAAGTCAAGCCCGTC
AAGAATCTGAAAGAAGACTTGCAGAAAAAGAAGAAGAATTTGAAGGTACACGAAAAAATCATCAACGAACACTCGAATCC
ATGCAAGCATCTCTTGAAGCTGAAGTTAGAGGACGAACTGAAGCAATGAAAATGAAAAAGAAGCTGGAGCATGATATTAA
TGAGCTTGAAGTTGGGTTAGACACGGCTAATAGACTGAAATCTGAACAAGAGAAAAATGCTAAGAAATATCAGCAACAAT
TAGCAGAAATGCAAGCTCAAGTTGAAGAACAGCATCATCAACGAGAGCAAGTTAGTGAACAGCTTGCGTTAACAGAACGA
AAATTAGGTATGATGCACGGAGAGCTGGAGGAAATGAAAAATATGTGTGATCATGCTGAAAAAAATAGAAAAGTCGCTGA
ATCTGAAAAGAATGAAGCCATTGATAAACTAAATGAGTTTTCAATTCAAAATTCTTCATTCATGGCAACTAAAAGAAAGA
TGGAAGCAGATTTAGCAGCAATGCAAGCTGACTTAGAAGAATCTACAAATGCAGCTAGACAAGCTAATGAACAATTGAAA
AAGGCAATATTTGATAATACTCGACTTTTCGATGAAATTAAACAAGAACAAGAACATGCTCAACAAGCTGAAAAAGCAAG
AAAGAATTTGGAGAGCCAGTTGAAAGAATTGCAATCAAAACTTGAAGAAGCAGAAGCAAATGTACTTAAAGGAGGCAAAA
AGGCTTTGAGTAAATTAGAGCAAAGAATAAGGGAATTGGAAGGCGAACTAGATGGTGAACAAAAACGACATGTTGAAACT
CAAAAGAATGCTAGAAAGTCAGATAGGCGACTGAAAGAAATTTCGTACCAGATCGACGAGGATAAAAAGAATCAAGATCG
AATGCAACAACTTATTGAAAGTTTACAAGCAAAAATAAAGACTTATAAACGACAAGTAGAAGAAGCTGAAGAAATCGCTG
CTGTGAATTTGGCAAAATATCGCAAAATTCAACAAGAGATTGAAGATTCTGAAGAGCGAGCTGATCAAGCTGAACAAGCT
TTACAAAAATTAAGAGCAAAGAATAGAAGCTCGGTATCTGTTAGTCGAGGAGCAAGTTCTGGACCTGGAACCAAAATTTC
TATAACTGCGACATCAATCCGCAGTGAGGACTAATCTGACAGAAAAAATTCATCATTAGGAAATATGCAATTAGTTAGCC
AATATTTCCCTTAGACTAGATTTTAAACCTTTAAGACATTTTTTTCAAATTAATAAAACGTTTTGA
>SMED30000153 ID=SMED30000153|Name=Netrin 1|organism=Schmidtea mediterranea sexual|type=transcript|length=1945bp
TTACTTCATGAAAATGCAAGAAACTATTAAAAAATAAAATTACTTATTTTAATCTTGAATGAATTCAACGAATATTTTGT
TGATTTTATTGAATATCATTTGGAAATTGACTATAATTAAAGGTTTTTCCTCGTGTTTTTCCGAGTCCGGCAGAGCAAAA
TTCTGTGAACCTGACTTTATCAATGTTGCACAAAATGCTACATTTGAAGTTACAAGTACATGCGGTCAGAAAGGATCAAC
AGACAGAATTTGCCGAAACTGGTACACATCTTCAGGCTTAAACGAAGGCAAATGTTTGAAGTGCGATGATAGAATTCCGG
AATTATCGAGAAATATATCTCTCGTATTCGATCGAAACATTCAAGGCCATGAAACGGTATGGGTTTCTGGACGTATTCCA
AAATTTAGTGGAATTCACTCCATTAATATCACGATTAATTTAAATAAGCTATATGAAGTATATTATATAATTGTCGAATT
TGCCGGTCAGCTTCCAGATTCTATGGTTTTATATAAAAGGGATAAAAATGGAAAAACTTGGTTACCTTGGAATTATTTTT
CGTCTTCTTGTAAGAAATCATTTAAAATGAAGAGAAGTAAACCTTTACACATTCAACAACGATCATTTAACAAGATAAGT
CCTGCTTGCTTTTCTTTCAAACACAGTGTCCAAAATATCACATCTAATATTAAAGTTCTTCCATTTAATCCGATGACAGG
CCGAACTTCTTTATTACCGAATTCCAGATTAATTAAAGAATGGGTCAGTGTTACTGATATTATGATATCATTACTAACTG
ATGGCTCTAAAAAACGAAATAAGTCACCATATGATCACTTGTCAATATCTGATATTACGATTGGTGCTAGAAGTCAGTGT
TATGGTCATGCTAAGAGTTTAATTAAAAAAAATGGCACCACAGTTTGTGACTGTCTACACAAGACAGCAGGACCGGATTG
TTCACAATGTCTTAAAACTCATAATGATCAACCCTGGAAAATAATTTTTCCCTCGGAACCAGCTGGGAATATTTGTGAAA
GATGTTTTTGTAATAACCATGCGGAAGAATGCAAATTCGATGAGAAAATTTATAAGAATTCTAGCGGGAAGCATGGAAGT
CGCTGTCTTCACTGTTTGCATCATACTACAGGTCCTCATTGCAACTCATGTTCCCCTGGTTACTATAGGAACCGTGCATT
CCCTATAAATCATCCTCAAGTCTGCCAAAAATGCGGTTGCCATCCTTACGGGTCTTTGCCCCAATCTTATTGTAATCCTG
ATAATGGGATCTGTCAATGCAAAATTGGTGTCGAGGGTAAATTATGTAACAGGTGTAAAAAAGGCTACCAACAAACAAAA
TCTTCAAGCACACCATGTGCTCCAGAGACGAAAAATAATCTCATGGCTACAGAAGAACGTTGCGGAAGTTGTGGGAAAAA
CCGTCCGAGAATTAGGTTTCGAAAATTCTGCAAAAAACACATAATGATTAAAATATCCATTCAATCAAAGGAGATCCACG
GTGAATGGACTCGATTCGGAGTTCGGGTGTTAAAGATCTGGAGAATCCAGCAATCATTAAAATCACAAGTCAACCTCTAT
GAATCTGAGTATTCAATATACCCAATATGGGTGGAAAATGCAGACATACGCTGTAAATGCCCGAATCTTCAACTTGGTCA
CACCTACCTCCTTTTGACTAAAATCAAATTAAAAAATTATCTCAATCAGCCAGAATTAATCGTTGATAGAAAGAGCAGTG
CACTTGATTGGAGTGACGACTGGATTCGAAGGCTGGACATGTTTAAAATGAGAGAGGAACAGGGTAAATGTGACCGCTAT
CAATATAAACGGAAACACTCGAAATTGATGAACTCTAGATGAAATGTTGAGTTGACCCTTAGATGTCTAGATATTTCATT
TTTACTTTTCATTCTCCATGTAAAA
>SMED30031287 ID=SMED30031287|Name=Cathepsin L|organism=Schmidtea mediterranea sexual|type=transcript|length=505bp
TTGGGCATTTTCAACGACTGAATCATTGAAAGGTCAAAACCAAAGTAAAACTGATAATTTATTCAGTTTTTCTGAGGAAC
AATTGGCTGATTGTAGTCAGGCATACGGTAATTTTTGTTGTGAGGGTGGTCTGATGGACTTTGGTTTCCAATACGTTATG
ATTAATAGAATTGAAGCTGAGGATGATTATCCATATTCCGGCATCTTGAGTTTCATAGGATAATAATTGCCGATGTAATG
GATTCAAAGCAGTCTGGGAATGTATGGAATACAATGACATCGAATCAGGACATGAACAAGATCTTGTAATTGCCATAGCG
ACAGGTGCACCAGTAAAACTATAATTCTGCGCAACTGCAAGTGTACTTGACATATGTTTAGAGACGGAGTTTATGATGAA
CCCGGATGTGCATTGGATAACCTAGACTATGGAGTATTTGCAATACGGTATGGAAATGAGTATGGCAATGACTATTGGCT
TGTCAAGAATAGTTGGAGAACACGT
>SMED30011264 ID=SMED30011264|Name=Macrophage-expressed gene 1 protein|organism=Schmidtea mediterranea sexual|type=transcript|length=2311bp
AGTCGCGATAGCACGCAACGTCGACATTGCGATGACGTAATCATTGTGAATCTCTACTACAATCGGATCATTCTAATCAC
GTGCATCCTAAAACAATATAAATCTTACAAACGGGAATCCAATTGAACATTTATTTACTCAAGCGTGAAACCAATGATAC
CGTGTAGACTTAATAGTATAATATTGTTTATATTTGTTTATGGAGTTCTATCGGTTTCATGTGAAATTGAACCTAACCGA
GATCAACCATCTTCAAGACGAGTTCATGAGTGTAAACTCACCAGCAACCAATATCCAGTGAATGTTCTTCCCGGATCTGG
GTGGGATAATCTTGAAAACAAAGAACTCGGTCAAGTTTTAGAAATTTCATATAAAGGATGTTTTGGATCGAATGATGGAA
GCTTTCTCATTCCAGACAATGTGGTGATCACCCCAATAAAGAAATCTATTGTCGAGTTAACTTCAGATATATTTGACCAT
TTCTCAAATGTCACCTCTACAACGGCGATGTCTATCAACGCCGATGCCAGTTACTCAACACCAAGCTACAAAATCAGTGG
ATCATTTTCATTTGAAAAAATAAGAATAAAAAACGATCAAATCAAAGAAAAGTCGTCGATTTCGAGATCCCAGTTAAGAC
ATGAATTATATGAAGTAGCGTTCCAACCACACTCAAAACTAGACGCAAAATTCAGAAACAGAATTCTAGATATTGGAGCG
TTATTGAAAGAAGGAAAAGACACAATTTATCCACAATATTTAGCGGATTTAATTGTCCGTGACTTCGGCACTCACGTCAT
TCGGTCGGTGGATGCGGGAGCGACTTTAATAAAGACTGACGTTCTGAAGTCGAAATACGTTAATAAATTCAAAGGAGATA
CAACAACCATAGCAGCCGCCGCATCGTTTAGTATGACAAAAGTTCTTGGGGTAACAATCAAAGGAAACTACTTCAGTGAC
ACTGCTAGCCTCGATGAATACAACAATAACAGCGCAAGCACTAAAGTTGAAACAGTTGGTGGAAGACCATTCGGTTCGAA
CTTCAAATTGGAAGATTGGGAAAAATCTCTGGATTCAAATTTGGTGGCCATTGATCGTCGAGGCAATCCATTATTCTACG
CAATAACACCAAGAAGTCTCCCGGAATTTGATGATAAAACCGTTAATAAAATTGTTACTCTTATCAAAAACGCTGTCAAC
AGATATTATGCAGCCAACACTAGAGTAGGTAAGTTAAACTTTAATAATAAAATACAACGTTGAGACTAGGGTGCATGAAT
ATGGAATCAGAAAATTTCAATGACGATGCAAATATTGATGGTGGAAATTGTGAAACTCCAAAATCATATTACACACTTGG
AGGAACATTTCAAACCTGCAAAGGGCCGGATTACCAATGTGAAAACGTAGCTGGCAAGAACCCCATAACGAATAGCTATG
CGTGTCCTGCAAATTACAAAACCGTTGAATTGGAAAGTGGATTGAGTAAATGCGAAAAGGTTTGCGTTAGGACTTTTTGG
TTTAGTTACAACTGTCATTACGAATGTTCCTATTACGCACTCTATTGGTGTGCTGCAACGAAAGCGACAACAAAATCAGA
AGGATCATTATTCGGTGGATTTTACTCAAATACATATCCGAACCCGATAACAAATGCCTATAATTGTCCGCAATATTATT
ATCCATCTCGATTTGGAATGGTCGGAACGATTTGTTTGGCATTCAATAGAGCAGAGACAATGGAACACCATTTGCCTTTC
GGAGGATTTTACAGCTGCAAGGTCGGAAATCCACTCTCAGACAAATTTGTAGATAATACCACACAGGATCCCAAATATTG
GTCGGAAGGTTGCGCAACTGGATACTCATCTCATGTGGCTATTGTCAACCGAGGCTGTCAAATTTCTTACTGTATCAAGA
ACAACGCGCTTAGCAAAAGGGAGCAACCTCCTCTTTTTAGACCCCCATTTGTTGATTTGGGATTCGGACCGAATGCGACC
ATCACTACTAGTTTAGAAACTCCAACTGGACTCAAACTCTCCGTCGACGAAAACGGGAAGTGGAGTGAAAACAAAAAATC
AAAATCATCCAATTATAATATTATTATATTTTCTGTAACAGCAGTAGCAATAATCACCATAATGATTATAATATTAATTC
TTGGAATTAAAATTCATAGAAAAACTCGAAGAATAACCGTTTTGCCAGAATATAATGGCAGCGATAGGAGCAATTCTACG
AGTTCTTTGAATAATCCAACAACATCGAATCGCGTTCAGCCATTCAAAATACTTATAGATGAGAATCAATA
>SMED30022992 ID=SMED30022992|Name=Myosin heavy chain|organism=Schmidtea mediterranea sexual|type=transcript|length=6157bp
ATATATATGCATCTCAAAATTGGAAATTAAATTATTAATTTTAATTTACTAACAAATTTTTGTTGTAAAATGGAAAATAT
TGATGCTAAAGACTTGCAATATTTACAAGTTGATAAAAGTTTAGTTTCTGACTCTGCTAATTTGTCTGATTGGGCTAACA
AAAAATTGGTTTGGATACCTGACGACAATGAAGGATTTATTTCAGGATCACTTATAGAAGAAAAAGGCGATGAAGCTACT
GTCAAACTGGAAAATGGCAAAACAATGAAATTACCAATTGAAAATGTTCAAAAAGTGAATCCACCAAAATTTTTTAAATC
TGAAGATATGGCAGATTTAACTTATTTGAATGAAGCATCAGTTCTATATAATTTAAAAGACAGATATTTTTCAGACTTAA
TATATACATATTCCGGATTATTTTGTGTCGTTGTAAATCCATACAAGCGCTTGCCTATTTATACAAATAATGTAATTGAG
TGGTACAAAGGTCGAAAACGTCACGAAAGACCGCCACATATTTACGCAGTTACTGATGTCGCATATCGAAATATGCTCCA
AGATAAAGAAAATCAATCAATTCTTTGCACTGGTGAATCCGGAGCAGGAAAAACTGAAAATACGAAAAAAGTTATTCAAT
ATTTGGCCTCAGTTGCAACTTCTTTGAAAAATCAAAAAACCACAGCCAGTAATGCATTGTCACAATTTTATGCTCACGAT
GTTAACATTGGAGAATTGGAGACCCAATTACTTCAGGCCAATCCGGTTTTAGAAGCTTTTGGAAATGCAAAGACTATCAA
AAACGATAATTCATCTCGTTTTGGTAAATTTATTCGAATCAATTTTGACACTTCTGGATTCATTTCCAGTGCAAACATAG
AAACATATTTATTGGAAAAAGCTAGAGTTATTAGACAGGCAGCTAATGAACGGTGCTTTCACGTGTTTTATCAGCTTCTT
CTTGGTGCAGACGACCGATTGAAAAAGGAATTAATCTTAGAAAATGTTTCTACATACAAACTTTTATCCAACGGCATGAT
AGCTGTACCAGATTACGATGAAAAGCAAATGTTTAAAGATACAGTCGAAGCTCTCGATATAATGGGGATTTCGAAAGACG
AACAAGAATCTATTTTCCGAGTTATAGCCGCAGTTTTACACATGGGAAATATCGAATTCAAACAAGAAAGAAGTTCGGAT
CAGGCTGCTTTACCAGACAATACAGTTGCTCAAAAGGTTGCTCATCTTCTCGGTTTACCGGTCACAGAAATGACCAAAGC
ACTGCTGAAACCAAAATTGAAAATTGGAAGAGAAGTTGTAGCAAAAGCTCAAACTAAAGAACAAGCCGAATTTTCCGTTG
AAGCTATAAGCAAATCAACATATGAAAGAATGTTTCGATGGCTGGTAATGAGAATAAATAGATCAATTGACAGAAATAGA
CAAAAAACTAATTTCATTGGCATTCTTGATATTGCTGGCTTTGAAATATTTGAAATAAACTCTTTTGAACAATTGTGTAT
AAACTATACCAACGAGAAATTGCAACAGCTTTTCAATCATACAATGTTTGTGCTAGAACAAGAAGAATACAGAAAAGAAA
ATATCAAATGGGAATTCATTGACTTTGGTCTTGATTTGCAGCCAACTATCGAACTAATTGAAAAGCCAATGGGTATTCTT
TCTTTGTTAGATGAAGAGTGCTTTTTCCCAAAAGCTACCACAAAGACATTTGTTGAAAAATTAATTAAAAACCAATCGAC
GCACCCTAAATTGAAAGCTACTGATTTTAGAGCAAAAGCAGATTTTGGGGTTATTCATTACGCGGGAAAAGTTGATTATG
TCTCCGATAATTGGCTGGTGAAAAATATGGATCCTCTTAATGAGAACGTTGTTAGTCTCTTACAAGAATCAAATGAAAGT
TTTGTCCAGACAATTTGGAAAGATGCAGAAAACATTATTGGCTTATCAACTACTACCGCCCAAGAATCTGCCTTTGGAGG
TGCAAAAACTACTCGCAAAGGAATGTTTAGAACCGTCGGTCAGCTTTACAAAGAATCATTAACGAAATTGATGGATGTTC
TGAATAATACCAATCCCAATTTTGTGAGATGTATCATTCCCAATCATGAAAAAAAATCTGGTAAAATCGATTCAAAATTG
GTTATTGATCAATTAAAATGTAATGGTGTGCTGGAGGGTATCAGAATATGCCGACAAGGATTTCCAAACCGAATTTTATT
CCAAGAGTTCAAACAACGTTATGGAATTCTTACACCAAATGTCATACTAAAAGGTTTTATGGACGGTCGAAAAGCAACAG
AATTAATGCTTGGAGAGTTGGATATTGATGCATCAAATTACAGAATTGGTCAGAGTAAAATATTTTTCAAAGCTGGAGTT
TTGGCTAGATTAGAGGAAGATCGAGATATTAAGTTGACTGAAATCATAGTCAAATTTCAATCTTATGCACGTGGATATTT
GGCAAGAAAAAATTTACAAAATCGATCGCAAAATCTCAATGCAATTAAAATTATTCAAAGAAATTGTAATGCATACTTAA
AATTGAGAAATTGGGCCTGGTGGAAATTATTCACACGTGTTAAGCCTCTATTAAGTGTTACAAGGCAAGAGGAAGTTGTG
GCTTTGAAGGAAGAAGAATTGAAAAAATCAAAAGAAACTCTCGAAAAGATGACTGCTGATTTCGAAACAACTAAGAAAAA
TTATGAATTTCTCATGGAAGAAAAAAATAAAATTCAGGAAGATTTGGAAAAGGAGCGTTTTGCATTGCAAGATATGGAAG
GGGAAAGAGAAAATTTGATCAAGCGATTAAAAGATATTGAATCAGATGCTCAAGAATGCGAGTCAAGAGAGATGGAAACA
AAAGACAAATGCAATAAATTGGAAATCGAGAAAAATAAATTACAAAATGAAATTTCACAGCTTAGCCAAAATTTAGAATC
AGAGGAACAGAACTTGCAAAAAGCTCAAGCAGACAAATTAAGTGCTGATAAACGAATTAAGGAACTGGAACAAAAAGTGG
CCGAATTAGAAGATAAACTTAACAAGTTAACTAAAGAGAAAAAGTCTCTAGAAGAGCGATTGGCTGATGTTATGTCTACT
TTGGCGGAAGAAGAAGAAAAGAGTAAACAATTAACGAAACTCAAATCCCGTCACGAAACAAGTATCACTGAATTAGAAGA
GAGATTGAGCAGAGAACAAGCAGCTCGACAAGATTTAGAAAAAACAAAGAGACGACTAGAAACTGAATTGTCTGAAAAAT
CTGATAATTTATCATCTCAAAGCCATACATATGAAGAAATTAGAATTACTCTTGAACGAGCAGAAACACAAATAATTCAA
ATGCAAGCTAAATTAGATGAAGAAACAACGGCAAAATCGTTGGCTCAGAGACAAATTCGAGACGCAGAAAACGTAATCAC
CGAAATCAAAGAAGATCTTGAAGCTGAAAAACGTTCAAAAGAGAGAGCTGAAAAAGCTAAGAGAGATTTAGCCGAAGAAA
TAGAATCTCTTAAAATGGAATTACTCGATACGGGAAATACATCAGAAGAACAGCAAAGCGTATTGCGCAAAAAAGAAGTT
GAATTACAAAATTTTAAAAAATCTTTTGAAGATGAAACAAAACGAACTGAACTTGAAATGCAAGAAATTCGTAAAAAAGC
CGCAGTAACGATTGAAAACCTCAACAATCAAATAGAAGCTGCTAACAAATCAAAACTTGTTGTTGAAAAGGCTAAATTAC
AGTTGCAAGAAGAACTTAATGATGCAAATAACGAATTAGCAAATTTAAGATCCTCCAAAGCTGACTCTGAAAAACTTAAG
AAGAACACAGAACTACAACTTTCAGAAATATCTCATCGCCTTTCAGAATCTGAAGAACACAAGCTAGATGCTGAATCAAA
ACTTAAAAAACAAACCGAGTTGGCTGACAAATTGGCTATGAATCTCGAAGAAATTGAAAGCAAATTGTCGCAATTAACTA
AATCTGAATTATCAGTTCGAAGTCAGTTAGCAGATTTACAAAAGGACTTTGAAGAAGAAACTCGATTAAAATTGAATGCT
CAAACTACATTAAGGCAGTTAGAAAGTCAATTTGCTTTATTAAAAGATCAATTAGAAGAGGAGCAACAATCAAAAGAAAA
TTTAGAAAGTCACGTACAAGCTCTACAGAGTCAAATGCAAGATGTCAAAAAGAAAGTTGAAGAAGATGCTCAACAGCTAT
TAGAATTTGATGAATCTCGTAAAAAAGTAATGCGTGAACGAGAAGAATTGCAGATTAGACTTGAAGAAGCAATCAATCAG
ACTGAAAGATCAGAAAAAGCTAAGCGAACAATTCAAGCCGAGCTTCAAGATGCATATCATGCCCTAGAAAGTCAGAAAAA
TGACCAAACTCAAAGTGATCGAAAACTTAAAAAAATGGAATCACAAGTCAATGAAACATTGAACTTGAATAAAAAATATT
TAGCTGAAAAGGATACATTAGAAAAAGAAAATCGTGAAAAAGAAACAAAAATCATGCAATTACAGAGAGAAATGAATGCA
AATGACGATAAACTTTCGGAATTGGAAAATGCAAAATCCTTACTGACTCGGCAATTGAATGAATTGGTTTCGAGCAAAGA
TGATGTTGGTAAAAATGTTGGCAGTTTAGAAAAGGCTAAAATTCAATTAGAAACCGATTTAGAGAATTGTAAAGTTACTA
TTGAAGAATTAGAAGAAGAAAATGCAACTCTTCAAATGAATAAAGAACGAGTTGAAATGCAATTAACTGCTACTAAAGCC
CAACTGGATAGGGAGCTAGCTTCCAAAGATGAAATGCACGATGAACAGTTCAGAATTATAAACAAAAAGCTTCGTGATGC
AGAAGCGGCACTTGAAGAAGAGCAGAAATCCAAAACAGCTATGGTTTCATTGAAAAAGAAAATAGAAAATGAGAACATGG
ATTTATTAAATCAATTAACAGAATCTGAGAAGGTCAAAGAAGAGCTGGCCAGACAGATTAAAAAATACCAAGGAAATTCT
AGCAATCTTCAAAGAGAAATGCAAGAACTTGTTGCTGCTAAAAATGATTATTTGAAGCAGATTAAAGAACTAGAAAAGCG
CTTACGTACTTCTGAAAGCAATGTAAACCAATTATCCGAAGACCTTTCGCAGTCTGAACGTGCTAAGAGAACCATTGCAT
CTGAAAGAGATGAAGCTTTAGAAGAAATTGCCACAATCACTATGGCTAGGGATACAGCTATTGCAGATAAAAAACGTATT
GAGAGTAGCATTTCAGCATTAAGCATGGATAATGAAGAATTGCAATTACTTTTGTCAGAATTAGAAGACAAAAATAAGAA
AATTATTTCTCAGTTAGAACAAGTTCAAAATGAATTAAATGTTGAACGACAAAATATAAGTAGATTAGAAAATCAAAAAT
CAACTGCCGAAAGACTGAATAAAGAATTGCATGATAAAATTGAAGAATTGGAAATGGAAGCTGGTAAAAAATATAAAGCT
ACAGTTGCTGCTCAACAAAGTAAAATCCAAAGTCTTGATTCTCAGCTGGACGCTAAAATTGAGGAAATAAACCAAGCTAA
TAGAAATAACAAGAAACTAGAAAGAAAGTTGAAAGAAATTATGGCTCTTATGGCTGATGAAACAAAGCGGGCTGACAATG
CAATCGAACAGATTGAAAAAGCCACTGGCAGATTTAAAAAAGCAGAGAGAAGTTTACAGATGGAACAAGAAAACAATTCT
GCTCTAAGTACACAAAATCGCCGATTACAACGAGATTTGGAAGAAGTAAATGAAACCAAAGAAGCACGGGACCAAGAAAT
TAAAATTCTTAAAACCAAAATCGACAGATTGGAAAAAAGAACACGCGCTGGGGGTCGACCTCTTAGAGGAGACGGTGGAT
CTGATGATGGGTTATCTGGAGATGGAGAATCCGTAACTGATTCTGCTTTAAAATCTGTTCAAGAAGACTAATTCAATAAG
TCCTAAAATGAACAATAAATTCTTGAATAATTAGTTAATACAAAAATTTTTGAATAATTTATATTTGGACTAACTTTTAA
TTCCTATTTGGTTTGCTCAATATATAATTGTTTTGATATATTTAATATTTCTATTTTATCAATTTGTGTACTTGGGG
>SMED30023593 ID=SMED30023593|Name=Netrin 1|organism=Schmidtea mediterranea sexual|type=transcript|length=1962bp
GATCTTGAAATTCTTTTTAAAATTTCTAATCTTATTCATTGGTCTTGTGGCCTTAAATGGAGAACTGCAGACCTGCTACA
GTGCCAATGGAGATCCTAAACCTTGTGAACCATTATTAATAAATTTGGCTCAGCCAATGCATATTGAAGCTAGCTCTACG
TGTGGAGAAAACGGAGCGGAAAGTATTTGTACAGAATGGTTTGACAAACTAGGAAAAGCTCATAAGTCCTGTGAGAAATG
CAATTCTTTAGATCCTAAATTATCAAAATCGTCAAAAATGGTGGTTGATTCACATATACGAGGCAAGGAGACTTGTTGGA
TTTCAGAAGGTGTTCATGAAAGTAGTTATCCAACTAGCGTTAATCTTACATTGGGTTTGAAAAAGAGGTCATTAATATCA
TTTATTACTGTGGAATTATGTGGTCACAACCCGGATGGCATTATTATTTATAAAATGGACAGTGAAACAGATGATTGGTC
TCCTTGGCAATATTTTGCTAAAGACTGTCAACTGACATTTAAGATGCCGGCTTTAACAACGGATACAGTTAATACTTTTT
CCGATATAAATTCATTGAAACCAGTTTGTTTTGATTTGGAAAAAAGTATGGACTATGGAGGAGATGAAAAACTATTCAGA
CTTATCCCATTCAATCCAAGACAAGGCAGAAGTTTTGGAGATGTATTATCTAAAAATGAACAAAACTGGTTGGCAGCTAC
AGAAATACGAATCAGTTATATTCTAACTCCTAAAGAACATTACGGAAATCTGCTGTTGAGAGAAAAGCGAAATGCAGAAA
ATCCTGTTTATCATTTTGGTTCATCTGATATAATGATTGGTGGGCAACCTTTATGTCATGGACATGCTTCGAGAATAATC
AGAACTTCGGATGATACTTATCAGTGTGAATGTCAGCACAGTACTGCTGGAAAGGATTGTGAAAAATGCCATGAAGAATT
CAATGATCAACCGTGGAGTTCAGCAACGATTCGAAACCCAGCGGTTTGTAAAAGATGCGAGTGTAACAATCATTCGATAA
AATGTGTATTCGATTTTAATGTTTTCCTGAAATCTGGTAGAATTCATGGCAGTTCTTGCATAGATTGTGGTGATAACACT
GTCGGCCCTCAGTGTGATCAATGCGCTCCCGGATATTTCCGGCAACATGCTTATCCTATTTACCATAAAAATGCTTGTAA
ACCTTGCTTATGTCATTCAGTTGGATCTCTTTCGGGTACACCCTGCGATCAAGTAAGTGGGAAATGTTTCTGTAAGCCTG
GAGTGACTGGGAAAACATGTACCCATTGTGCACCGGGTTATAAACAAACAAATTCCCTAAAGAATCCTTGCAGAAAGATA
TCAAAATTGCGTCATTTAGTGAAATCAAATTCGAGTTGCGATTTTTGTCAAAGGAAAAGTAAAAGAATTAATTTTAAGAG
ATTTTGCAGCAAGGAAACAAATTTCAAAGTAAATATCAAGAGTAGAGAATCAAGGGGTCAGTGGATTCGGTTCATTTCTG
AAATCGAACAGGTCTGGACAAGAAATCAACAAGATCCCCGAGTATCCCAACTTAATCTAGAATCTAGTGAAGGGACTTCG
ATGTGGGTTCATAAAGATCTCGTGGACTGCGGTTGCGTTGATTTTGAATTCGGTCATTCATATTTAGTCATGGCCAATGT
GAAAACAACGGAATTTATGGGCCAAACTGAACTTATTTTAAACCAAAGTGATGTAATTGAGGATTGGAAGGAGACATGGG
AGGAGCAATTTTCTCAGTTTTATACCCAGGATAAAATGGGAAAATGCAATCGGTTTACTCGACAAAAGCGACACGTCAAG
AGGCGTAATTTTGCCTCAACCTATAACCAATATGCTATCTATTCCTGATCATTTGGACCCGGGATTAATTTAATTAACAT
TTTATCTAGCAGCTGAATGTGATGTCTTATTGTATTTATAAA
>SMED30001362 ID=SMED30001362|Name=SMEDWI-3|organism=Schmidtea mediterranea sexual|type=transcript|length=3093bp
AATTAAGAGACAGCCATTTATGTTTATTTTTAAAATAAATAGTATATATTTAAGTTTCGAATTTTCAGCTACAATTTAAT
AGTACATAATTATACGATGTCAGGAAGTAGTGGAATAGGTAGAGGCCGCAGTCGTGGGCTGTTGATGCAAAAGTTTCTGA
ATAAAGATGTTCTTGTTCCTTCTGTTGAATCTTTAGAAGACAAAGCTCTTAATAAGCTAGGAATTCCACTTCCTGGGTCT
ACCGTAGAAAAAAATACAGAGTCAAGTTCTATATCAAGTGGAGATTCAAGAAGTAATCCCAGTGGAGATTCAAGAAATAA
CATAAAACTACAAGATAGTGATATTGAGAATCGCAATATTACCATTGTTACGCGACCATTATCCTGCATAGGCAGGGGTC
GGGGTTTAAGCAATCCCTCAAGTTTAACTACGTCATCGGGTAAATCGGATAAAATTACTGAAAACGAAGAACCTGGTCAA
ATTAAAAAGTTTGTAGGTCGCGGTAGAGGATTGTTGAATTCTCAGAAAGAATGTTCAAACTCTACTCCATCTGAAGTTTC
AAATGAATTGAAACAAATGAAAATTTCAAATGATGATAAAATGACGGTTTCTTCAGAAGCAAAGTCACAATTTGAGAACA
TTGAAAAACCTATTAGTAAATTTCGTCGACGCGAATATCCAACTCAAATAAAAGAACCATGTAATACAAGAAATGATTCA
TCGCCATCTTTAACTTTAAGTGCTAATTACGTTAAAGTTAGGACTACACAACCCCATATATATCAATACCATGTTTCCTT
TGCACCTCCGATAGATTCAAGGTTGATGAGAATTAAAATAGTTCAAGGGTTATCAGAATCAGATTTAGGGGTTGTCAAAG
AAGCAAGAGCTTTTGACGGTATGAATTTATACATTCCTCAACTTTTAAAAAATAAAGAGACAATAATTAAAGTAAACAAA
CCGACTGATAAGACTGTCGTGGATGTTAAAGTAGTTTTTACTAACAATGTTAATTTTAGTGAATGTCCTATGGTTTATAA
TGTTCTTTTTAAAAGAATTGAAAATTCCCTCAGAATGGTTAAAATTGGTAGGGATTATTTCTACCCTGAAAAAAAGATAG
TACTTGACCGTAGAAGGATGGAAATATGGCCGGGATATGTAACAAGTATCCAAAATTTTGACGGTGGTTTACTGTTACAA
TGCGATGTGTCACACAAAGTTATTCGAAATGATAGTGTGTATGACATAATGATGGAAATTAATAAAACTGTTAACAATAA
AGGTCAAATGCAAACTGCTGCGATTAATCAACTATTGGGTCAAATTGTGTTAACTCCTCACAATAACCGAAATTATAGAA
TTACTGATATAGATTGGGCTAAAAATTGTTTAAGTGAATTCGATAAAGGAGGCGAAAAAATTAGCTACCGGGATTATTTT
AAGAACACGTATGGGCTACAAATTCGTGATCTAGAGCAGCCTATGATAGTTAGTAAATCTAATAGCAGATCTGGTAAAAA
CCGAGGTCCCAAAGGATCAAAAGAAGTGGATGGTGGATTGGTCTATTTAATTCCAGAATTGTGTATGCTAACTGGTTTGA
CAGATGACATGATTAAAGATTTTCGTTTGATGAGAGAATTACACGAGCATTGTCGAGTTACTCCCAAGAAAAGACACGAA
GCCTTACTGGAATTCGTGGATAACATATATAGCTGTGAGGAAGCTAAGAAACTTTTAGGGTATTGGGGTATAACGATTGA
AAAGGACACTGTCAACATAAATGCTTGTAAAATGAATCCAGAAATGATATATTTTGGAAATGAAGCTTCTGTTAGTGCTG
GGGAACAAGCTGAATTTAAACAAGCCTTGGCACATAATAAAGTTATAGGTGGTATTCGTATTGAAAATTGGATATTAATT
TCTCCAAAAAGTTTACTGACAAAAGCAAATGGTCTGTTACAGGCTTTAATGAGCAAATCTCCTAGAGTTGGAGTTATGTT
TGGAAAACCCAAAATAGTTGAAATGAACAATGATCGAACAGAAGAGTATTTAAAAGAATTAAAGAGAAATGTGGCTCCTG
GTGTGCAGTTAGTAGTTACAATTTTATCTGCTGTTAGAGAAGATCGATACAATGCAATAAAAAAATTTTGTTATGTGGAT
TGTCCTGTTCCAAGTCAAGTGGTATTAGCCCAAACATTGAAAGAAGGGCCTAAATTAAATAGTGTGGCAGTTAATATAGC
CCTTCAAATAAACGCAAAATTAGGTGGAGAGCTGTGGGCTGTCAAAATACCTATTAAGAAGTTTATGGTTGTTGGACTTG
ATGTTTGGCATGATACTAAAGGGAGAAGTAGATCAGTTGGAGCCGTAGTTGGTTCAACTAATGCGCTATGCACAAGGTGG
TTTTCGAAATCGCATTTGCAAGAACAAGATAAAGAAATTATGTACGTATTACAGTCGTGTATGTTAAGCCTTTTAAAGGC
TTATTTTGAAGAAAATAATTTTTTGCCTGAGACTATCTTTATGTATAGGGATGGTGTTAGTGATGGTCAGTTAGGATATG
TTCAAAAAACTGAAATTGAACAATTCTTTAAAGTTTTTGAATCGTTTAGTGCTGATTATAAACCTAATATGGTATATAAT
GTTGTTCAAAAGAGAATAAATACTAGGCTCTATGTAAGTGATCCGAAAAATAAAGGACAAATAAATAACCCCAATCCTGG
TACAATTGTCGACCATACTGTTACGAGGGCTAACCTTTATGATTTTTTTCTTGTTTCTCAATCGGTTAGGCAGGGAACTG
TAACTCCGACGCATTACGTTGTTTTATGTGACAATTCTAAATACACTCCGCATCAGGTTCAGTTGATGGCTTATAAAACA
TGTCATATATATTACAATTGGCCGGGAACGGTTCGAGTACCAGCACCTTGTATGTATGCTCATAAATTGGCATATATGGT
TGGTCAGAATTTGAAAGCTGAACCTAGTAATCTTTTATGTGACAGACTTTTTTATTTGTAACATATGTGTTTCTATTTGT
ATTATTGTTAAACTGCATTTCGTAAAATAAATTAATTTCGTTATAAAAAAAAA
>SMED30016824 ID=SMED30016824|Name=Cathepsin L|organism=Schmidtea mediterranea sexual|type=transcript|length=375bp
AAATGTATTTATGATAAATCAAAAGTTGTTGCAAATTTCACTTCGTTTACTGTTATCGAGCCAGGAAACGAATCTGACTT
GGCAGCCGCCGTGGCCACCGTTGGACCTGTGTCCGTATGCATCGATGCCGATCATCCAGGTTTCCAACATTATAAAACTG
GTATTTACAATAACCCCAAATGTTCATTTTTATGCTTAAATCATGCCGTTTTAATCGTTGGTTATGGATCACTCAATGGA
GATAACTTTTGGATTGTGAAGAACAGTTGGGGACCAGATTGGGGAATAGAAGGATACATACTCATGTCAAAGGACAAGGA
CAACCAATGTGGAATTGCTAGTCTAGCCGCTTATCCAGTGGTCTAATATGCATGC
>SMED30000139 ID=SMED30000139|Name=Macrophage-expressed gene 1 protein|organism=Schmidtea mediterranea sexual|type=transcript|length=2115bp
GTTTGATAAAAGTTATAATTTTATTATTAAATAGTGAAAATGACAATTTTTATGTTGGGTCGAGTAATTTTATTCCTCTC
AATCGGATCCATTTTGGCTGTGCTGGCTGAGGAGAAAGAGCTAAATATTACTCATCCCCACAAAAATATTGCCACTTCGA
ATTTGCCATCAGCCAAATTTGTACATGGATGCCATTTGACCAACAATCAGTATCCATTCAATGTGCTTCCTGGATCTGGC
TGGGATAATTTGTCAAACCGCGAGTTGGGACAAGTTCTAAAAGTAGCCTATTCTCGTTGCAAATCATCCAATGATGGAAC
CTTTCTCATTCCGGACAAAACAAAAATTACGGCGTTTATGAAATCTAGAGTTCAAATGGATTCAGATATTGTAACACACT
TTTCCAAATATTCCTCCTTGACTTCGAAATCAGTAAATCTTGAAGTCAGTTCTGCAACTCCGAATGTTCAGATCAGCGGT
TCATTTTCTTATGAAAAGAAAGACATCAAGGACAATCAAATGAAAGAAAAATCCGCCTTTTCTCAAACTCAGTTAAGATA
TGAATTGTATCGAGTATCCTTCGAACCGGATAGTGAATTGAATGACAGATTTCGTGAAAGGATCCTTGATCTTGGAGTAC
TTATAGAGAATAAAAAGGGTTCAGCTCATGCTCACTATCTCGCTGATTTGATTGTCAGAGATTTCGGAACTCATGTTATT
AAATCTGTTGATGCTGGAGCAATTCTTTCAAAAATTGATGTCCTAAAAGGTGACTATCTTTCAAAACACGAGGGCTCTCA
ACATTCCGTGACATTAGCTGCGTCAGTTAGTATGAGAGAAACACTTGGAATTAAAATAAGTGGTAATTACAAAACCGGTC
ATTCGGAAATGGAAGAGTACAACAAAAATTGCGTAAGCACTCGAATCTCTACAATCGGTGGTGGACCTTTCGGTTCGAAT
TTCACGCTAACAAAATGGGAAAAAACTCTTTCAACAAATCTTGTGGCCATTGACCGTATTGGGGAACCGCTTAATCACAT
TATTACCAAACGTAACGTACCGGAGTTTGACGATAAAATTGTGTATGAAATCGGAAACCATATCAAGGAAGCAATAAGCC
GATATTATTCTGCTAATACTCGAACTGGATGCATGGACATTATGTCAAAGAACTTTGATGAAAATGCAAATGCCGACAGT
GGATTATGTAATAACATTGAAAATGGAAATTTCTTTGGGGGTTTATACCAAAAATGCAGTTTAGACGATGACTCTCTCTG
TGAGAATATAATACAAAAAAATGCCGCTACTGGGGATTACTCGTGTCCAAATGGGTTCACTCCAGTCAAACTATATTCAT
TCAAGTATAAATGTGAAGGTGATAACTGTCCTATTTATGAAACCTTCTGGTGTGTCTCTAACGGTAAATCGTCAAAAGGA
TATGAAATTTTTTTCGGAGGAATCTACACAACAAATATTCAGAATCCGTTTTATAGGGCTGACAGTTGTCCGCAATACTT
CTTTCCTGCTAATGTTGGAATTAACGGTAAAGTCTGCATTTCAATGGATAAAGAACTGGGATTTAAAAATAGTTTACCAT
TTGGAGGATTTTACAGCTGTTCTATGGGAAATCCATTAGCTCAAAAGAAAGGATTTAATTCAACTGTAAATGGAAGTTAT
TTTCCAAAAGCTTGTGGAAGTGGCTATTCTTCTCATTTTGCTCTCCTAGATGGTGTCTGTCAGTTGTCCTATTGTGTTAG
TACTAATGCACTGACAAAGGGATATGACCTACCGTTGTACAGACCCCCATTTATTGATATTAAAACGGAATCAGAAGTTG
TCAACATCAACGCATTGAAAACGTCGGCTGGAATAAAAATGATTCTTTCTGAAAATGGATCTTGGGAAACAATGATTCCT
GAATTCTATTCCACGTACAAATATGCAATAATAACAGTGCTATTTCTCGTGCTATTATCCTTGCTTTTCCTGATGGCAGT
TTTAGCTTGGAAGGGTTTTAGACAATTTCTTGGAAGGATTTTCACTAAAATACCTTTAGTGAATAAAAAACAATCAAACC
ATAAACCGATTCGCGTACAAATAAAGGTGACCAGA
>SMED30015696 ID=SMED30015696|Name=Macrophage-expressed gene 1 protein|organism=Schmidtea mediterranea sexual|type=transcript|length=2665bp
AGTCGCGATAGCACGCAACGTCGACATTGCGATGACGTAATCATTGTGAATCTCTACTACAATCGGATCATTCTAATCAC
GTGCATCCTAAAACAATATAAATCTTACAAACGGGAATCCAATTGAACATTTATTTACTCAAGCGTGAAACCAATGATAC
CGTGTAGACTTAATAGTATAATATTGTTTATATTTGTTTATGGAGTTCTATCGGTTTCATGTGAAATTGAACCTAACCGA
GATCAACCATCTTCAAGACGAGTTCATGAGTGTAAACTCACCAGCAACCAATATCCAGTGAATGTTCTTCCCGGATCTGG
GTGGGATAATCTTGAAAACAAAGAACTCGGTCAAGTTTTAGAAATTTCATATAAAGGATGTTTTGGATCGAATGATGGAA
GCTTTCTCATTCCAGACAATGTGGTGATCACCCCAATAAAGAAATCTATTGTCGAGTTAACTTCAGATATATTTGACCAT
TTCTCAAATGTCACCTCTACAACGGCGATGTCTATCAACGCCGATGCCAGTTACTCAACACCAAGCTACAAAATCAGTGG
ATCATTTTCATTTGAAAAAATAAGAATAAAAAACGATCAAATCAAAGAAAAGTCGTCGATTTCGAGATCCCAGTTAAGAC
ATGAATTATATGAAGTAGCGTTCCAACCACACTCAAAACTAGACGCAAAATTCAGAAACAGAATTCTAGATATTGGAGCG
TTATTGAAAGAAGGAAAAGACACAATTTATCCACAATATTTAGCGGATTTAATTGTCCGTGACTTCGGCACTCACGTCAT
TCGGTCGGTGGATGCGGGAGCGACACTAATAAAGACTGATGTTTTAAAATCGAATTATGTCAAAGGTTTTAAAGGGGATA
CAACTAAGATTGCAGCTGCGGCATCATTCAGTATGACAACAATGCTTTCAGTCAACCTCAAAGGAACCTATGTTGGTGAT
ACCACGAGTCTCAATGAATTCAATAGTAATAGTGCAAGTACAAAAGTCGAAACTTTTGGTGGTATACCGTTTGGTTCGAA
TTTTAAGTTGGAAGATTGGGAGAAAACCCTTGAAACGAATTTGGTAGCAATTGATCGCAAAGGTGAACCGTTGTCCTATG
CTATCACACCAAAAAATCTTCCAGAATTCGATGAACAAGCAGTTTTCAAAATAGCTACGCTCATCAAAGAAGCAGTCAAT
AGATATTATGCAGCCAACACGAGAGTGGGATGTATGAATATGGAGTCAGAAAATTTCAATGACGACGCAAATGTTGACGG
TGGAAACTGCGAACCTCCTAATACATTTTACAGTTTTGGAGGAACTTTTCAAACCTGCTCAGGCCCTGATTATCAGTGTA
TGGATGTCGCGGCAAAAAACCCAGTGACCAATAGTTATGCTTGTCCTGATAACTATGATACCATTGAATTACAAAGTGGA
TTGAATAAATGCATAAATTATTGCACCAAATACTTATTTTCCACTCAATGTCATTACGAGTGTTCCCAATATGCATTGTA
TTGGTGTTCTTCTACAAACAAAACGAAAATATCAGAAGGCGCACTTTTCGGGGGATTCTACACTAATACATTTCCGAATC
CACTTACAAATTCCTATAGTTGTCCACAATACTATGTAGCGACAAGATTCGGCGTAAACGCCAAGGTTTGCACTGCACTC
AATAAGGCAGAGGCTATGGAACACCATTTGCCGTTTGGAGGATTTTATAGTTGCAAAGTTGGAAATCCTCTACTAGATGA
TCCTCTCTACAACATTACACAGGATCCCAAATATTGGCCTCAAGGTTGTTCAACCGGATATTCATCACATGTAGCTATTG
TCGACAGAGCTTGTCAAATTTCCTATTGCGTTAAAAATAACGCCCTTAGCATGAGAGAGCAACCGCCTCTCTACCGACCA
CCTTTTATTGATTTGGGATTTGGACCCAATATGACAGTAATTACCTCTCTGGACACTCCAATGGGGGTCCGATTATCTGT
TGATGAAAACGGGAAATGGAGTCATAATCACCAATACAAATTATCCAATTACAATATTATAATAATATCAGTCATTATGG
CGGCAGGAGCCAGCATTGCAATTATTATTATAATTCTTGGAGTTCAAAGATTCAGCAGAAACCGAAAAGGTCAAGTCTCA
CCGGAATCTCATGAATTTGATAGCAGATATGATTCTACTAAAGATGTTGCCAATCTTGCGTTACCCGGCGAGGTTCCACC
GTTTCGTGTTATAATTGACGGAGCGAAATGATGCTTGGAACCTTTTATTATTTCTGTTGAATCTGATCTGTAAATGTTTT
CAGAATATTTTTCTAATAAAATCGTTCTGAAATTTTAAAGTGATCCTTGATGCTATTAAATGATGGACTTTATTGTACCC
CGAATTGGGTCACCCTTGTTCACGCAAATTCGTGCTTAGGATATTTGTACGGGCTTATGCAACTTTTTAAATTTTTATTT
TTCCTTATTTTTATTAGCAATCCGTTTTATTTATAACGACAAATGCTTGATGTTTGAATCAAATTTTTATTATGTTTCTT
AATGAAACATAGTGAACAATGTGCCACTAATTTCATAACCAAGGTCTAATCAAATAGTTTGGGTAGTGAAAGCAATATTA
CAGAGTTTAGGTTTGTCATTCATAT
>SMED30010115 ID=SMED30010115|Name=ribonucleoside diphosphate reductase subunit M2-2|organism=Schmidtea mediterranea sexual|type=transcript|length=1127bp
GTTTTTAATGAACTGAATCAGGTTTAATGCAAAATAGTCATTTTAATTTCTAGATAAATTAACAGTAATTCTTTCGAGAT
TAGATATTTATTATGGAGATTTTATTAACAGAAGATAAAAGTCGAATAGGATTTCTTAATATACAATATGAAGATATATG
GGAAATGTTTAATAAAGCTCGATCTTCATTTTGGACTGTAAATGAAGTTAATTTATCAAAAGATTTTGCTCACTGGAAAA
AAATGAATGACGATGAACGGTATTTCATTACTCACGTTCTTGCATTTTTTGCTGCCAGTGATGGAATTGTCAATGAAAAT
CTCGTCGAAAGATTCATTCAAGAAGTGAAAATTCCCGAAGCTGAGGCATTTTACAGTTTTCAAATTGCAATTGAAAACAT
TCATTCAGAAATGTATTCTCTATTAATCGATTCTTATGTTACTGATACAGAAACAAAGAATAAATTATTTAATGCCATCG
AAACTATGCCATGCGTTCAGAAAAAAGCTCAATGGGCATTGCGTTGGATTAGCGATAAAAAATCCACTTTTGGAACTAGA
GTTGTTGCATTTGCTGCTGTCGAGGGAATATTTTTCAGTGGTAGTTTTGCATCGATATTTTGGCTTAAAAAGCGTGGATT
AATGCCCGGATTATGTTTCAGCAACGAATTAATTAGCCGAGATGAAGGACTTCATTGCGATTTTGCTTGTTTGATGTTTA
AATATTTAAGTAGCAAACCGTCACCTGAACAAGTTTTGGATTTGATCAAAGAAGCTGTCGGAATCGAGAAGGAATTTCTC
ACCGACGCATTGCCGGTTTCATTGATTGGCATGAACGCTGAATCGATGTGTGAGTATATTGAATTTGTGGCCGACCGGCT
GCTGGTTGAATTGGGCTGTGACAAGTTCTGGAACACAGAGAACCCGTTGGATTTCATGGATAATATTTGCATCGATGGGA
AAACTAACTTCTTTGAGAAACGAGTCGGTGAATATTCCATGTCGAGCGTTGAGGACAAACACAAATTTGATGATGTCAAT
TTCGATATTGATTTTTGAGGGATTCCGTTTTTTGGCGCCAGTGTGCATTTAATGTGTTTTGGTTGTAATTGGCTATTTTT
GTTTGTT
>SMED30034696 ID=SMED30034696|Name=Macrophage-expressed gene 1 protein|organism=Schmidtea mediterranea sexual|type=transcript|length=2009bp
AGCGAAAATATGACTCGTTTTTCTGTTGCCAGTGCAACATTTATTTGGTTTTTATTTTTCGGTTCCACACTAGCGGATGA
AGCAGTCATTGAGCCGGATAACGTAATTCAGAACTCGAATTTTCCATCCAAACGTGTTGTAAATGATTGCCACCTGACTA
ATGATCAGTATGCATTCAATGTCTTGCCGGGGTCCGGTTGGGATAATCTTGCCAATCAAGAGTTAGGCCAGATTCTGAAA
TTATCCTATAAAAACTGCAAATCTTCCAATGATGCCAGCTTCCTAATTCCCGATGAGGTTTCAATGTCACCAGAAATGTT
GTCTAATGTTCAATTGGATTCCGACCTAGTCTCACATTTTTCAAAATACGATTCAATGACTTCCAAGTCTATTAACATTG
AAGCAAACGCCGCAACGCCAAATTATCAAGTCAGTGGATCGTTTTCTTTTGAAAAGAAAGATGTTAAGAAAAATCAAATG
AAAGAAAAATCCGAAATTTCTCATACGCAACTGAGATACGAACTTCACCACCTTGCCTTTCAGCCAGACAGTGAATTGGA
TCCTACCTTTCTCAATAGGGTCATTGACATGGGAATAGTTTTGGAAAAAGAAAGTAAAAATACTTACTCAGAGTTTTGGC
TGATTTGATCGTAAGAGACTACGGAACTCATGTAATACGGTCTGTTGTCGCTGGAGCTATTCTTTCTAAAACTGATGTTC
TCAAAGGCAATTTTATGTCCAAATCCAGGGGAAATTCTATAAATGTTGCGGCCTCTGCCGCTGTAAGCATGAGAATGGCT
GTGGGAATTAAGATTGGAGGAGGTTATGGCAATTCTGAAACCAATGCCACGGAATATGAAAAGAATTGCGCAAGTTCAAG
AATTAACACCTTTGGTGGAGGACCATATGGTTCTAATTTCAAATTAGAAGATTGGGAAAAGACCCTTCCTACTAACCTTG
TTACTATAGATCGTACTGGTGATCCTTTATATTACGTAATAACAAAAAGAAACGTACCGTATTTCAGTGACAAAACAATA
TTTAACATCAACACTTTGATTAAAAATGCAATCAACAGGTATTACTCAGCTAATTCTCGCCCTGGATGCATGGATATTAA
ATCTGATAATTTTGATGAAAATGCTAACATTGACAATGGCATATGCCAAAATCCAGACGATAATTTCAAATTCGGAGGAG
TTTATCAATTTTGTTATGGGCTGAGTGCTAAATGTAAGGAACTGAACCAACTCAACCCAATCACTGGAACATCCTCATGT
CCAGATGGATTTGACCCTGTTGAGCTATTTTATGTATATAGTAATGAGAATAACGATGGTATGGTCATCAACACGTTTTG
GTGTGCTCCTAATCAAAAATCAACTCATTCAAATGGATATTTATTTGGCGGGATTTTTACCGATGCATATCCGAACCCTG
TTACTGGATCTTCTAGTTGTCCACAATATTTTCTATCTTTGAAATTTGGACGTACCGCAAAGATTTGCGTTTCACAGGAT
ATCGAACTTGGAATGAAAAATAGTCTGCCTTTTGGAGGGTTTTTCAGTTGTTCCATGGGGAATCCCTTGACACAGAAGAA
GACGACTAATTCTCCCGCTGACTATTGGACCCAATCATGCGCACCCGGTTATTCTGCACATTTGGCGGTCATCGAAATTA
ATTGTCAGATTTCATATTGTATCAAGAGTAACGCATTAAAAGGCAAAGAAGAAATTCCTCTTTATCGGCCACCATTCATT
AATTTAGACGATGGAATTAAGTATAATTCTATAAAGAAATTTCAAACTACTTCCGGGATAGAATTGAAACTTTCCAAGGA
CGGAATGTGGAATAAAGTGAAATATGTTCCTTTCGCAAGATCTAAATTTGGTTTAATTATTTTTTCCGAGGTTACCCTAA
TCTTGGTAATTATTGTTGTGGTTTTAATGGTTTGTTGGTTTAAATCCCGTTAAAAGAAAAATTGTTTTAAATAAAGACAG
ATCGGAAGA
>SMED30025388 ID=SMED30025388|Name=Myosin heavy chain|organism=Schmidtea mediterranea sexual|type=transcript|length=6089bp
TTTTTTTTCGAGTTTGTGCTAGAAATTAATTTCAGTTTGAAATTTTTTGCCTATTTCGAGTTCTTTGAAAACAATCTGAA
AACATGAACCCATCTGATCCCGATTTTCAATATCTCGGTGTCGACCGAAAAGCTTTAATGAAGCAGTATGGGGATTCCTT
TGACAGCAAGAAAAACTGTTGGATTCCTGACGAGAAAGAAGCCTTCATTGCGGCGACAATTGAAGATGCGTCTGGAGATG
TCTATACGGTCAAAACCGATAAAAGTGAAACCAAGACTGTTAAAAAGGACGAAATCGAACAAATGAATCCACCGAAATTT
TTGATGATTGAAGACATGGCAAATTTAACCTTTCTAAACGATGCCAGCGTTCTTGATAATTTAAGACAAAGATATTACAA
GAATCTGATTTATACCTACTCTGGACTGTTTTGTGTGACAATAAATCCATACAAAAGATTTCCAATTTACACCCCGCAGG
TGATTTCAAAATACAAAGGTAAACGACGAACTGAAATGCCTCCACATATTTTCTCGATATCCGACAATGCTTACACCAAC
ATGTTGGCGGACCGAGATAATCAATCTGTCTTAATTACTGGTGAAAGTGGAGCTGGGAAAACTGAAAATACAAAAAAAGT
TATCACTTATTTTGCACACGTTGCAGCTGCTACTAAAAAAGAAGAGGATGATACTGGAACTGGTCAAAAGAGAAAGGGAA
CATTGGAAGACCAAATCGTGCAAGCAAATCCTGTTTTAGAAGCATACGGTAACGCAAAAACTGTGCGAAACAACAATTCA
TCCAGATTTGGTAAATTCATTCGTATTCATTTCGGTACAAGTGGAAAAATAGCTGGAGCTGATATTGAATTTTACTTACT
CGAAAAATCGAGAGTAAATTCTCAACAGAAAGGTGAAAGAAACTATCACATTTTCTATCAAATTTTATCGAACGGAGGAA
AACCATTTCATGAAAAATTACTCATCTCTCCTGATCCAGCTTTGTATTCCTTCATTAATCAAGGTGAATTAACAATCGAT
GGAGTCGACGATGAAGAAGAAATGAAAATAACTGATGAAGCTTTCGACATTTTGGGATTTTCTGCCGAAGAAAAAATGAG
CTTGTTCAAGTGTACTTGCTCAATTTTAAACATGGGCGAAATGAAATTTAAACAAAGGCCAAGAGAAGAACAAGCCGAAG
CCGACGGTACTGCTGAAGCTGAGAAAGTGGCTTTTCTGTTGGGTGTCAATGCAAAAGATTTAATGCAATCGATTCTGAAA
CCGAAAGTTAAAGTTGGTAATGAATATGTCACAAAGGGGCAGAATAAAGATCAAGTTCTTTATTCGGTTGGTGCGCTAGC
AAAATCTCTTTACAACAGAATGTTTGCGTGGTTGGTTTTGAGAGTTAACAAAACCCTCGACACTAAAGTCAAGCGACAAT
TTTTCATCGGTGTTTTGGATATTGCCGGTTTCGAAATTTTCAATTTCAATGGATTTGAACAAATTTGTATTAATTACACC
AATGAAAGATTGCAGCAATTTTTTAACCATCACATGTTCGTGTTGGAACAGGAAGAGTACAAGAAGGAAGCGATCGATTG
GGAGTTCATTGATTTTGGAATGGATCTTCAAGCGTGCATTGAGCTCATTGAAAAGCCAATGGGTATTCTGTCAATCTTAG
AAGAAGAGTGCATGTTTCCGAAAGCATCTGATATGACATTTAAAGCAAAATTGTATGATAATCATCTCGGAAAAAGTCCC
AATTTTGGAAAGCCAAAACCTCCAAAACCCGGACAGGCTGAAGCTCACTTCGAATTGCATCACTATGCTGGCAGTGTGCC
ATACAATGTTACTGGTTGGCTGGAAAAGAATAAAGATCCTCTCAACGAAACTGTGATAAATCTACTTGCGGCAAGTAAAG
AGGCATTAGTTTCGAGTTTGTTTGCTCCGGCTGAAGATCCAAATAATACGGGTAAACGGAAAAAGGGTGGGGCCATGCAG
ACCATTTCTTCAACTCATAGGGAATCCTTGAATAAATTGATGAAAAATTTACAAAGTACCAGTCCACATTTTATCCGATG
CATTGTTCCAAACGAATTCAAACAACCTGGTGTAATCGACGCTCATTTGGTATTACATCAATTGCATTGTAACGGTGTCC
TTGAAGGCATCAGAATCTGTCGAAAGGGTTTCCCAAACAGAATGATTTATTCTGAATTTAAACAAAGATATTCTATCTTG
GCTCCAAATGCAATTCCACAGGGATTTGTTGAAGGCAAACAAGTAACGAGCAAAATTCTGGAAGCTGTTCAATTGGACAC
CAATCTTTACCGCCTGGGAAATACTAAGGTATTTTTCAAAGCTGGAACTTTGGCTGATTTAGAAGACATGCGTGATGAAA
AATTATCTTCACTGATTTCATTGTTTCAAGCTCAAATACGCGGATATTTGATGCGAAGACAATATAAAAAATTACAAGAT
CAGAGAGTTGCTCTATCTATCATTCAAAGAAATATTCGTAAATATCTTCTTCTGAGAACATGGCCATGGTGGAAATTGTA
TACCAAAGTTAAGCCACTTTTGAACATTGCTCGTCAAGAGGAAGAAATGAAAAAGGCCGCTGAAGAATTGACAAAATTAA
AGGAAGAATTCGAAAAAGTTGACAAATTCAAAAAAGAACTTGAAGAACAAAACGTCAAATTGTTGGAAGCTAAAAACGAT
TTGTTCTTACAATTACAAACCGAACAAGACAGTTTAGCCGATGCCGAGGAAAAGGTTTCAAAATTGGTTATGCAAAAAGC
CGATATGGAAGGAAGAATTAAAGAATTGGAAGATCACCTTCTAGAAGAAGAAGATAATGCTGCTGGACTGGAAGATATCA
AGAAGAAAATGCAGGGTGAGATTGAAGAGTTGAAAAAAGATGTTGTCGATTTGGAATCTTCATTACAAAAGGCTGAACAA
GAGAAAACCGCGAAAGATCAACAGATAAAATCGTTACAAGATCAAATGGCAAAACAAGAAGAAGAGATGAACAAATTGAA
AAAGGAGAAAAAGGCCGCCGATGAAGTGCAAAAGAAAACCGAAGAATCTCTACTAGCGGAAGAAGAGAAAGTCAAAAATC
TAAACAAAGCTAAAGCCAAATTGGAACAGACCATTGATGAGATGGAAGAAAACCTCTCGCGAGAACAGAAAATCCGAGCT
GACGTAGAAAAAGTTAAACGAAAAATCGAAACTGAACTGAAACAAACTCAAGAGACCGTAGACGATTTGGAAAGAGTAAA
ACGAGAATTAGAGGAACAATTGAAGCGCAAGGAAATGGAATTAGTCAATGCTTCATCGAAAATTGAAGACGAATCGGGAT
TAGTTGCCCAGCTTCAGAGGAAAATCAAAGAACTTCAAGCTCGAATTCAAGAATTGGAAGAAGATCTGGAAGCAGAAAGA
CAAGCCCGTGCAAAAGCAGAGAAAAGCCGACATCAACTCGAAGGCGAGTTGGAAGAATTGAGTGATCGACTGGAAGAACA
AGGAGGCGCAACCTCAGCTCAAATGGATCTCAATAAGAAGCGAGAGGCAGAATTAATGAAATTAAAGAGAGATTTAGAAG
AATCGAATATGCAACACGATCAGGTTATCGCACAAGCTCGAAAGAAGCAACAGGATGTTGTCAATGAATTCTCAGAACAG
CTCGATCAACTGCAGAAAGTCAAAGCTAAAGTTGAAAAAGAAAAGAATGAGATGAAAGAAGATTTAAATGACCTACAAGG
ACAGCTGGAATCATTAAATAAAGCCAAGGCTAACAGCGACAAACGAATCAAAGAAATCGAAGCTCAAAATTCCGAATTAC
AGGGCAAATTGGAAGAGGCAAATAGACATATCAATGATGCCAACAATACATCTGGAAAGAATCAACAAACCAATGCGGAA
CTCCAAGCACGTTTAGAAGAAGCCGAATCGCAAATCAATCAGCTGGGTAAAGTCAAGCAACAAATGCAGACTCAACTCGA
AGAAGCCAGGCAAATCGCTGAGGAAGAGTCGAGAGCAAAGGCTAAATTGAGTTCTGATGTTCGCAATCTCAATGCCGATT
TAGACAATCTCAGGGAGGCACTGGAAGAGGAAAATGAGAACAAAAGTGACATACAGAGACAGCTAGTTAAAGCTCAGGGA
GAAATGCAACAATTGAAGGCTAAATTCGAAGGCTCGGGATCCATTCGTTCAGAAGAGTTGGATGAAGCCAAGCGAAAGTT
TATGACACGAATCCATGAGCTGGAAGAGGAAGCTGAAACCGCCAAATCGAAAGCCGGACAACTAGAAAAAGTTAAAGCTC
GACTCCAAGGAGAAATCGAAGATATGCTGGTCGACGTTGACAGAGCGAATACACTCGCATCTCAGTTGGAAAAGAAGCAG
AAAACATTCGATAAAGTCGTGTCAGAATGGCAGCAGAAATATGCTGAATCTCAGGCGGAATTGGAAGCATCCCAGAGAGA
AAGCAGGACGATTTCAGCTGAGGTGTTTCGAATGAAAGCACAAATCGAAGAAAGTCAAGAGCAGTTGGAAAGCGTTAAAC
GGGAAAACAAAAATCTAGCAGATGAAATCCACGATTTGACAGAGCAAATCGGAGAAGGAGGTCGCAGTGTTCACGAAATC
GACAAAGCAAGGAAACGATTGGAAATGGAAAAGGAAGAATTGCAACATGCGTTAGAAGAAGCCGAACAAGCTTTGGAACA
GGAAGAAGCCAAGTCTCAACGATCACAACTGGAAATGAGCCAGATCAGACAAGAAATCGACAGGCGATTGGCTGAGAAGG
AAGAGGAATTCGAGGCCACTCGAGTCAATCATCAAAGAGCAATGGAGTCAATGGAAGCCTCACTTGAAGCCGAATCCAGA
GGCAGGGCTGAAGCAACGAAGATGAAAAAGAAACTGGAGCACGATATCGGTGAACTGGAAGTTGCAGTGGATACCGCCAA
CAGAATCCGATCGGAGCAAGAAAAGAACGCAAAGAAATTCCAGCAGCAAGTTCAAGAATTGCTATCAATGGTGGAGGATG
AGAAACACCAGAAGGATCAGATACGAGAGCAAACGACGATGACAGAAAGGAAATTGACAATGGTCCTGGCAGAGCTTGAG
GAGGTTCGTGGCTCATTGGAAAATTCCGAGAGAAACAGGAAAAATACCGAGAATGAGAAAATGGAAATAACTGATCGATT
GAATGAGTTGAGTGTCCAGAGCTCCGGTTTCATGGCTACCAAGCGGAAATTGGAAGCTGATTTGGCTGCTATGCAGTCCG
ATTTAGAGGAGGCCGCTACTGAAGCCCGACAAGCCAATGAACAAGCCAAGAAGGCTGTTGCTGACAGTGCGAGATTGTTT
GATGAAATCCGGCAAGAGCAGGAACACGCACAGCAAATCGATAAGATCAAGAAACAACTCGAGGCCCAAAATAAAGAATT
GCAAGCCAAATTAGATGAATCGGAGAATAATGCTTTGAAAGGTGGCAAAAAAGCCTTGAGCAAACTAGAGCAACGCGTTC
GTGAACTTGAAGCCGAATTAGACGGAGAACAAAAAAGACACGTTGAAAGTCAAAAGAACTCCAGAAAAGTTGAAAGACGG
CTTAAGGAGCTTGGATTACAAACCGACGAAGATAAGAAGAACCAAGAAAGATTGCAAGATCTGGTGGAGAAGCTTCAAGG
TAAAATCAAAACCTACAAACGCCAGGTGGAAGAGGCGGAAGAAATTGCTGCTGTAAACTTAGCAAAATACAGAAAGATCC
AACAGGAAATCGAAGATTCCGAAGAAAGAGCTGATCAAGCTGAACAAGCGTTGCAGAAGCTGAGGACCAAAAACCGCAGC
TCCGTTTCGACAGCGAGAGGCGGTAGCATGGCTCCCGGCATGACCACAGTGAAATCGACATCACTCTCTGTTTCGGCTTC
GAGCCGACGACAAGGTTCTAGTGTTTCCAGAGAGAGCTAAGTCAGAAATTCTAGTAACACAACGATAAATTATTAGGATT
TACTTAATTGACATTATTTGCATGTTATCAATTATTGGACACAATTTAGTGATGCTTTTATTTAATAAAATTTACATTTT
GAAAAAAAA
>SMED30018602 ID=SMED30018602|Name=ATP dependent RNA helicase DDX5|organism=Schmidtea mediterranea sexual|type=transcript|length=607bp
TATTCAAAAGATAAACGATTGTTAGAAGTTGTTAGCAAAACTCCACGGGACAAAATGATTGTGTTCGCAGAAACGAAAAG
GAAAGCTGATATGATTTGTTCTTTCCTACGTTCAAAACAATTCAGAGTTGGTGCAATGCATGGAGATAAGTCACAGAGCG
ACAGAGAATATATTCTTAATGGGTTTCGAGGTGAAAATTTAAATATTCTCGTGGCAACAGATGTAGCAAGCAGGGGTTTA
GATGTTGATGATATTAAAGTTGTAATCAATTACGACTTCCCTATTCAATTTGAAGATTACATTCATAGAATCGGTAGGAC
AGGCCGACGAGGAAAATCCGGTGTGTCTTATACATTTTTCAATAGAAAGAATTCGAAAAGTGCTCGTCAATTGGTGAAAG
TGATGGAAGAAGCTAATCAATCAGTTTCAGATAGTTTGTATGATCTTGCATATATACCAAGAAGTGAGCGTAGACAAAAT
TTCAATCGTCCGCCAATGCCAGTGCCGCAAGTAAATAATTATCAAGTACCTCCGCCTCAATATAATGGTGGAAATAACAT
GTATCCGCAACCAGATTTATATATGCAAAACGGAGCAGCACATCACA
>SMED30035691 ID=SMED30035691|Name=Macrophage-expressed gene 1 protein|organism=Schmidtea mediterranea sexual|type=transcript|length=2718bp
TATATTTTAGAGATTATGATTATAAAAATTTAAAGTGCCGACAGTTAATAAAAACAATTAATGCTTACAACGTAAGGTCA
TCATGATGATGTAATCATTCGGATGTTCTATGACGTTCGGATAACCCTTATCACGTGGATCCTAAACAAAATATATCAAT
CAACTAGGAATTACAATATACATTTGTTTGTTCAACAGTGAAACCAGTGATACTTTGTATGTTTAATAATATAATAATAA
TTATATTTGTCTGTGGAGTTCTATCTTTTAAATGTAAAAGCGAATCAAACAGAGACCTCCCGTCAGAGAGGCAAGTCCAT
AAATGTAAACTTACTAACAAACAATACGCATTCGATATCCTCCCTGGATCTGGATGGGATAATCTTGAAAACAAGGAACT
CGGTCAAGTTTTAGAAATTTCATACAAGGAATGTTTGGCTTCGAATGATGGCAGCTTTCTCATTCCTGACAACTCGGTGA
TTATTCCGATCAAAAGGTCAACTGTCGAATTGGCATCTGACATATTTGATCATTTCTCTAATTTCACATCTGCCACGTCC
AAATCCATAAATGCCGATGCAAGTTACGCCACACCAAGCTATCAGATTAGTGGCTCGTTTTCATGGGAAAAAAAGGATGT
AAAAAATGACCAAATCAAAGAAAAGTCTTTGGTTTCTCGGGTACAATTGAGACACGAATTTTACACCGTTTCATTTCAGC
CAGACTCGAAATTTGATCGTCAATTCAAGAACAGAATTCTAGATATTGGAGCGTTATTGAAAGAAGGAAAAGACACAATT
TATCCACAATATTTAGCGGATTTAATTGTCCGTGACTTCGGCACTCACGTCATTCGGTCGGTGGATGCGGGAGCGACACT
AATAAAGACTGATGTTTTAAAATCGAATTATGTCAAAGGTTTTAAAGGGGATACAACTAAGATTGCAGCTGCGGCATCAT
TCAGTATGACAACAATGCTTTCAGTCAACCTCAAAGGAACCTATGTTGGTGATACCACGAGTCTCAATGAATTCAATAGT
AATAGTGCAAGTACAAAAGTCGAAACTTTTGGTGGTATACCGTTTGGTTCGAATTTTAAGTTGGAAGATTGGGAGAAAAC
CCTTGAAACGAATTTGGTAGCAATTGATCGCAAAGGTGAACCGTTGTCCTATGCTATTACATCAAGAACTCTTCCAGAAT
TTGACGAGAAAATTATTTACAAAATCGCTACTCTCATCAAAGAAGCAGTCAATAGATATTATGCAGCCAACACGAGAGTG
GGATGTATGAATATGGAGTCAGAAAATTTCAATGACGACGCAAATGTTGACGGTGGAAACTGCGAACCTCCTAATACATT
TTACAGTTTTGGAGGAACTTTTCAAACCTGCTCAGGCCCTGATTATCAGTGTATGGATGTCGCGGCAAAAAACCCAGTGA
CCAATAGTTATGCTTGTCCTGATAACTATGATACCATTGAATTACAAAGTGGATTGAATAAATGCATAAATTATTGCACC
AAATACTTATTTTCCACTCAATGTCATTACGAGTGTTCCCAATATGCATTGTATTGGTGTTCTTCTACAAACAAAACGAA
AATATCAGAAGGCGCACTTTTCGGGGGATTCTACACTAATACATTTCCGAATCCACTTACAAATTCCTATAGTTGTCCAC
AATACTATGTAGCGACAAGATTCGGCGTAAACGCCAAGGTTTGCACTGCACTCAATAAGGCAGAGGCTATGGAACACCAT
TTGCCGTTTGGAGGATTTTATAGTTGCAAAGTTGGAAATCCTCTACTAGATGATCCTCTCTACAACATTACACAGGATCC
CAAATATTGGCCTCAAGGTTGTTCAACCGGATATTCATCACATGTAGCTATTGTCGACAGAGCTTGTCAAATTTCCTATT
GCGTTAAAAATAACGCCCTTAGCATGAGAGAGCAACCGCCTCTCTACCGACCACCTTTTATTGATTTGGGATTTGGACCC
AATATGACAGTAATTACCTCTCTGGACACTCCAATGGGGGTCCGATTATCTGTTGATGAAAACGGGAAATGGAGTCATAA
TCACCAATACAAATTATCCAATTACAATATTATAATAATATCAGTCATTATGGCGGCAGGAGCCAGCATTGCAATTATTA
TTATAATTCTTGGAGTTCAAAGATTCAGCAGAAACCGAAAAGGTCAAGTCTCACCGGAATCTCATGAATTTGATAGCAGA
TATGATTCTACTAAAGATGTTGCCAATCTTGCGTTACCCGGCGAGGTTCCACCGTTTCGTGTTATAATTGACGGAGCGAA
ATGATGCTTGGAACCTTTTATTATTTCTGTTGAATCTGATCTGTAAATGTTTTCAGAATATTTTTCTAATAAAATCGTTC
TGAAATTTTAAAGTGATCCTTGATGCTATTAAATGATGGACTTTATTGTACCCCGAATTGGGTCACCCTTGTTCACGCAA
ATTCGTGCTTAGGATATTTGTACGGGCTTATGCAACTTTTTAAATTTTTATTTTTCCTTATTTTTATTAGCAATCCGTTT
TATTTATAACGACAAATGCTTGATGTTTGAATCAAATTTTTATTATGTTTCTTAATGAAACATAGTGAACAATGTGCCAC
TAATTTCATAACCAAGGTCTAATCAAATAGTTTGGGTAGTGAAAGCAATATTACAGAGTTTAGGTTTGTCATTCATAT
>SMED30033415 ID=SMED30033415|Name=Cathepsin L|organism=Schmidtea mediterranea sexual|type=transcript|length=703bp
AGAACAAAAGGATACGTAACTCCAGTGAAGTATCAAAAAAAATGTAAGTCCTGTTATTCCTTTTCAACTGCAGGATCTCT
TGAAGGTCAATATTTTCGGAAAACAGGCAAATTGGTTAATTTTTCTGCACAGCAATTAGTTGACTGCAGTAGTTCTTTTG
GAAATGAAGGATGCCGAGGAGGATTTGTAGAAACGTCCTACAGCTATGTGATGGAATACGGCATTCAGTTAGAAAGTGAT
TACCCGTATGAAGGAAAATTTAGACAGTGTAAATACAATAAATCTCAAGTCGTAGGAAAATGCAGTGGATTTGTTCGCAT
AGAAGAAGGGAATGAAACTGATTTGGCCATCGCGGTTTCAACGGTTGGACCTCTGTCCGTTTCAATAAATGTTCAAAAGG
ATTTCATGGATTACAAAAGTGGCATTTACAATAATGCGAATTGTCTGTTTTATTCATTAAATCATGCTCTTCTACTTGTC
GGTTATGGATCAGAAAACGGAGTCAACTATTGGCTTTTGAAAAATCAATGGGGAACTGCTTGGGGTGAAGAAGGATACGC
CAAAATGGTAAAAGACGCGAGAAATCACTGTGGAATCGCTACAGCTGCCAATTATCCCTTAATATAAAATAGACTGATAT
TAATAAAAATGTTTTGTTATAAATTTTAGAATTAAAAAAAGTAATAATAATAATAATAAATTG
>SMED30027646 ID=SMED30027646|Name=Transcription factor BTF3|organism=Schmidtea mediterranea sexual|type=transcript|length=541bp
AAATTAATTTGTTTAAATAAATTTCTTTAGAATCACGCAATGGACGGTTTTAAAATTAACACTGAAAAATTAGAAAAACT
TAGTAAGTTGTCGGAAAATGTCCGAACAGGTGGTAAAGGTACAATGAGACGAAAGAAGAAGGTCATTGTCAAGAGTTCAG
CAGCAGATGATAAGAAATTAGAGGCAACCTTCAAATCATTGTCAATGAGCCCGGTTCCAGTTGAAGAAGTAAACATGTTC
AAGGATGATGGAACTGTTATTAATTTCGTTAATCCAAAAGTCAATGTGTCACTTCCATGCAACACTCTCTCAGTTACTGG
AAAATCCGAAATTAAGCAATTGACCGAAATACTTCCAAAAGTTTGGCAACAACTTGGCCCACTTAAAGGAGATCTTTTCA
AGAAAATTGCACAAAACATGGAAAATCAAGCAACTGTCGAAGATATTGATGATGAAGACGATGACGTTCCAACTCTCGTC
ACAGACTTCGATCAACAAAGCAAATTGGAATAACTTTTTAATATCAATAAATTTTTTGGAC
>SMED30016085 ID=SMED30016085|Name=Macrophage-expressed gene 1 protein|organism=Schmidtea mediterranea sexual|type=transcript|length=1970bp
TTACAATATAGAAAATGCTTTGCTTTCCTCTGGGAGGGTTGATTCTACTGATATCCATAGCATTTAGCTTAGCTGACATC
AATCTACCTCAATATCAAAGAGGATCAAACACGAATTTCCCATCGAGACGAATGGCCCATGGGTGCCAGTTGACAAATAA
TGAATATCCATTCAATGTGTTGCCAGGATCAGGTTGGGATAACCTCTCTAATCGGGAGCAAGCACAAGTGTTAAAAATAT
CATATGACAAATGTAAAGCCTCAAATGACGGAAGATTTCTAATTCCTGATCAAACGAAATTAGTGCCAAAAATGAAGTCA
ATAGTGCAGTTAGATTCAAATTTGATTTCACATTTTTCAAAATATCCGTCGACTACTTCTAAATCTATTAATGCGGAAGC
AAGTGGAGAAACGAAGAAATATTCAATCAGTGGATCTTTTTCCAATGAAAGGGAAGACGTAAAACGAAATCAGTTAAATT
CTAAATCCTCAATATCCCAATCTCAATTGAGATACGATCTTTACAAAATTTCTCATATCGTAGACGCTGAACTTGATACA
AAGTTTCACCAAAGAATCCTAAACATTGGGGTGTTGATTGAAACAAAAAAGGATTCTTTATATTCAGCATATCTCGCTGA
TTTAATCGTCAGAGAATTTGGAACTCATGTCATTACATCGGTGGATGCTGGAGCCGTTTTGTCGAAAATTGACATCTTGA
AACAAAAATATGTCTTAGATAGTCAAGTACAAAATAGTGACGTATCAGTTGCAGCATCATTCAGTTTGGGACAAATTTGT
GGATTTAAATTCGGTTCAAATTTTAAAAATGATGACACAAAAATTGAGGAATATAACAATAATTGTGCCAGTACGAGAAT
TAAAACAATTGGCGGTGGACCTTTCGGCTCGGATTTCTCTTTAGAAGAATGGGAAAAAACCCTTGACACAAACTTGGTGG
CTATAGATAGAATCGGAGAGCCTTTGTATAACGTAATTACAAAGCATAATATTCCAGAATTTAGTGACAAAACGATTTAC
GAAATAAACTTACTTGTCAAAACTGCTATTGAGAAATATTATTCCACAAACACTCGATTAGGATGCATGGATTTTAATTC
ACATAGCTTTGATGAAAATGCCAATGTCAACTCCGGTTTGTGCAATGAAACGGGAAATACTTATCCCTTTGGTGGGGTTT
ATCAAACCTGCCAGAATTCAGACAAATGTGACACATATAATCAAAAGAATCCCGCCACCGGAGACTATAGCTGTCCAAAG
GCATTTAATCCTATCAAACTGTTTGAACTTGACAATCCAAGTATCACTTTTGTTTGGTGTGCTCCACGAAGCTCTTCAAG
CGCAATAAATGGATATCTATTTGGCGGAATGTATACTGAACAATATCCGAATCTTGTTACTAATAATCACAACTGTCCGC
AATATTACTTTCCTGTCCAGTATGGAATCAGTGGGAAAATATGCATAACTCAAGATAAAGAACTCGGCGGCAAAAATAGT
TTGCCATTCGGAGGATTCTACAGCTGCAAGACTGGAAATCCATTAGCTTTGGCCATTTACAATGGAACTCAGACAAACTT
TCCTCTGACCTGCGGTTCGGGTTATTCTACTCATTTAGCGTCTATTTTTAACAACTGCCATTATTCGTATTGCGTTAAAA
ATAATGCTATAAAAACGAAAGCTGCGACTCCACTTTACAGGCCTCCGTTTGTTGATTTGCAAATGGATTCGAATATTGAA
GTCGTCAGTGAACTCACTACTTCAGCAGGAATGAAGCTGGTTTTAGCCGAAGATGGAATATGGAAGGAAGAAATCCTGAG
AACTTACATGGTCGAGTATATAATGATAGCAGTAACTGTTGCGTTGCTGACTCTAATTGCTATATATATCAAGTTCAAAT
GTTTCGTTGGAAAGAAGTTAAATCCTACTCAAGTTTCAGCAGATCGGAAG
>SMED30025427 ID=SMED30025427|Name=Translationally controlled tumor protein|organism=Schmidtea mediterranea sexual|type=transcript|length=570bp
AAGTAATACTTTTATCATTTAATTGAAAAAATGCTCATTTACAAGGATATTGTTGCCGGCGATGAGTTGTGCTCTGATAC
ATTCCCAATGACGATTGTCGACGATGTCGTTTACGAATTCAACGGAAGAAATATTACTATTAAAAACGGTATTTCTGACG
CCCTCTTTGGTGGAAATGCTTCAGAGGAAGCTGCTGGCGATGATGACGTTGCTGATTCGGACGAGACAGCAATTGACATT
GTATACAGCAACCGATTAGTCGAAATGAGCTTTGACAAGAAAGGTTACACGAGTTACATCAAAGGTTACATGAAAAGTAT
TTTGGAAAGATTGAATGCCACTAATAAAGAACGAGCTGAAATCTTCAAATCCAAAGTCGGTGCCTATGTTACAAAAATCA
TCAAAAATTTTGATGAATATCGTTTCTACTGTGGAGAGTCAATGAACCCTGACGGAATGATCATTCCATGCAATTACAGA
GAAGACCAAATTACCCCATACTTTGTTCTCTTCAAAGATGGCCTCGAAGAAGAAAAATGCTAGATTTCTGTTAAATAAAT
TTAGTGGAAC
>SMED30023322 ID=SMED30023322|Name=Cathepsin L|organism=Schmidtea mediterranea sexual|type=transcript|length=613bp
ACGCATTCTCTACGACAGGCGCGTTAGAAGGTCAACTATTCAGAAAAACTGGAAATTTGATAAGTTTATCTGAACAACAG
ATTGTTGATTGTGATAGTTTTGATTATGCATGTAACGGTGGATTTGTGACTGATACTTTAACTTATATTAAACAAAATGG
AGGAATTGAATCTGAGCAAGATTATCCATATGTGTCAGGAAGGACGAAGAAACCTAATAAAAAATGTTATTTCAAAAAAG
AAAAAATAGTGGGAACAGATACTGGTTTCGTATCTTTACCTCCAGGAGATGAAGAAGCTCTGAGAGAAGCTCTAGTAACA
ATTGGGCCAATTTCAATAGCTTTTTCCGCGTCGCCTTTGTTATCAGGATATAAGAATGGGATATATCATGATTACATGTG
TGCTGATGTTCGCATCAATCATGCAGTTTTACTAGTAGGATATGGAACGGATGAAGAATCAGGAACACCTTATTGGATAA
TAAAAAACAGTTGGGGTCCCGCATGGGGAGAAAATGGATATTTTCGAATGGAAAGAAATAAAAACATGTGCAACATGGCA
TACAATTCATACTATCCATTGGTTTGATTGAACGACTTCTTATGAGTGTGTGT
>SMED30005748 ID=SMED30005748|Name=HNF4|organism=Schmidtea mediterranea sexual|type=transcript|length=3066bp
GAAGAATAACAAAGAAATTTATTGCCAAACTAAAATACCCGGCCTATTTCATCAAAAAAAAAACAATCAAAAACACAAAT
CGCTACGAAAACTTGTATAAAAGATTCACATTAAAAGTCATCAATATTCCTCTTTTTTAAAGACTGATGATTCAGATTCT
TGGATCCGAATGGGGTGATCTGTTAGGAAGTGAGCTGTTGACGAGATAGCTGAATAGTTTGATCTGGAATTGTGCTGTTG
GTTCGCTTCCGATGATCTGTTGGCGATTGATTGAATTTGGCAGTTTTCTATCGGCGAGTTTGGAAGCGACTTGGTATAGG
CCACAAGAAGCGACGGATCAACGACATTTTGAGTGGAATTCACAGAAAGAGCTGGGTGGATTAGGTCATCTGAATTGGAA
GAATTTGAGCAGTTATGATTTGCGTCACTGTTATAGGCGTATTTTGACTGGTTGATCATGGGGAACCCTTGATCAAGGTT
GACCAAGAAATGGCTGGTATCCTGTGATAAATTGGAATGCTGATAAAGATTGATGGAATCATTGACGCATGAAAACAGAG
AAGGAACGCCGACTCTTTCATTTTTCAGCATCATCCCATCGCTGTTGATTATCATACCGAAGTTGCTATTGTCCTGGGAA
TTTTGCTGCATTATTTCAGTTCCTTGATTAACAAAAGCTCGATATATATTTCCTGAAGGCATCGACGTAGAACTTGGCAA
ATATGCACTCCATATTTTTGAGGCCGCTTCAGGTGATATGTTTTGGTTGTTGATATTGAGTAGGACACTGTCTGGCATGA
AAGGGGGAATTCCGTAGCAGTCCTGGTAAGTATATTTCGGTGAGGAAATCCTGTGGGAGTTAATTATATCCATGGAATCG
TTTTGATTATTGTTATTGTTCCCATCTGTTTCTGGATCTATGCTGAATACCCCTGGTGGATTGTCACCCAATAATGTTTC
AGATAACAAGCTATCAATCTCGGCTAATCCTGTCATTTTCATGAACTCGACCTTCTTTACCATCAGCTGGGTGACCAATC
TCAGATCAGGAATTGTCAGCAGCAATTCCCCGAATCTACCCGGCTTGTGGTATTGCTTGTCATTCATGTGGTTCATCAGG
TCCATTTGTATTTGGTATCGGCATCGTCTCACGAACTCCTTGCCAGACTCGCTGACTCTCGGATCAAAAAATACGATCGC
TTTCAAACAAGCGAATTCTGCGTCATCCAATTGCAATTCTCGCAGCGGAATGAACAGGTCGTCAAGGATGTGACTGGCTA
TTTCAGCGAAATGTTTGTCACTGGAGTTCCTACTGATGATAAGATTGTTTCCCAATAATAGCACATCGTCTTCGTCCAAT
TGAAACGACCGCCTTATTACTCCAAGAATTAGAATTTCTCCCGCGTGTGCTTTTAAAAGGGAAATTTGATCGCTCAAGTT
TAACTGTGAAAAACAAGGCAAAGACTTTGCCCAATTCACTAGAAGAAATAACTGATTTTTCATGGACTCGCAAACATCAG
GCACATCGGCATATCTGTTCATGTATTCCTGAGGATTCGGGGGCTTTTGTATTGCAACCCTTTGCTCGGCCTGCATCAAC
TGAGAAATGGAGAGAATGACATTCGGAGGAATATCATCAAACGACGATCTTCGAGTACTGATCCTATCTCGTTCATTTTG
AACCGCTGCTCGCTTCATTCCTACAAGAATGCATTTCTTTAGCCGACAATAACGGCATTGGTTTCGTTTATCCTTGTCCA
TTTTACAATTCCGATTGTACTGGCAAGTGTAGTTATTCTTCTTCCTCACTGACCTCCGAAAAAATCCTTTACAACCGTCA
CACGAGAAAGCCCCATAATGTTTTCCTGTAGCTTTGTCACTGCAAATGAGACAGAGTTGATTGTTTTCCATTCCTTCTGG
TCCCGACGCGTTAGACGCATAGACTTCATGTTGTTGCGAGGACACTGAAACGAAAACTATTTGATAATATTTGTACAAAA
CAACTGACAGATGGTTTCGGAAAAGGTTCAATGTTCAGCCATAGTTCAGCGAAAGGTCAAATTCCTGAACTACTATTTCA
AGGTATAAATTTGAATATATTGGTTTTGCCTCGTGCTAACAAAGTCCAGAAATATAATAAGCAAAGTCTGAATCGTGTAC
TTTGGATTTCTAGCCAGGGGCACACAATAACCTATATTTAATAGCAGTTCATTGCGTTCACTCGTTATTTCTCCGACTGA
ACTGCATTGCAGGAGGATCCGCAACCATGGAGATTTCATAATTATTATCAAAATGATAATAATAATAATTATTATGAGGT
ATTGTAAAAGAAAACGTAACTCACTTTCACTTTCCAATGGATATTGCATCGAGGAAGCGTTGCTGGAAATGGAAATCGGA
GCTGAATTGCTAGTCAATAGTTGATAGGAGAATGCTGGATTGAGGTTCGGATGCTGGGTAGACGTCATATAGAATTGGCT
CGACGGATTCGGTAAAATTATGGTGAAGTTTACAATCGAAGCTCAGATATAACGTATTATAATGAAGTATTATTAATAAA
CTGCAAGTAACGTGCGCGCATTCTGAAGTGGTTGATTGAGAATAATCAGTTCTTATTCGACAAATTCAGATAAAAAAGGC
ACCACTCGAATTGTTCAGCTGCGTCATGTGAATGCCGGAACGTTGTGCGTTGGCTGTTGGCAGATCAGCGCTCCGTGTTC
ACTGCAATCCGGATACACCAGACCACTTGAAATCGTTGCGAATTCAGCGTGTCGAGTGCATTGTGCGAGATCAGTATCGG
TGTGTATGTGTGTCTGCCTGTGTGCGTGTGGATGTGTGTGCTTGTCGAGTCGGTTGTGTGTTTATGAGTGTAACTGTGTC
GATTTGGTCGAATGCAGTAGCAGATAGCTGAAATGCGATTGTTCCTATTTTTCATTTAATATTGTTTTATGTTTTGTTTC
TATTTTTATAATTTAATTTATCAGTTTTTGTTTAGATTATGCAATCATTGTTTAATTTATTTTTGTCAATTTAAAATTTA
TTATTAATTAAATTATTATAATTATT
>SMED30002168 ID=SMED30002168|Name=Spondin-1|organism=Schmidtea mediterranea sexual|type=transcript|length=2693bp
CAATTGCTTAATAAAATTTTCCTTTCAATAATTACCGTAAAAGTTGATGAAATTAATTTTGAATATTTTATCCACTTTTT
TCATATTGAAATTTGATTTTGCTGAATGTTTCGGATTTTGTGACATTCAATTTCCGAAATCATACGTTGCCTTTTCCACT
AGATTCAATCTCTATACGAATCATGAAAAAGAAAAACTGACCAAAATCACCTCGTACATTCCTGATAAAACCTACGACAT
AACTGTGTCATTGAATGATAAAGCTATGCACGAAAAGTTGACAATAAAAGAAATATACGTCTTAGTCTCAAATGTTCACA
CGGAAATGGAAGAATTGTCGGAACATGACTGGAAAAACGCCGGTGAAGTGGTTTTTAATGTAACTCACGCATCCAAAACA
GAATATTGCCCCCAAAGAGCTTACGTTCAGTATTTATATACCCAACCCCAACATGTCACATTCTATTGGATAGCACCTAA
AATACCGAATTTTTGTATTAGAATAACAGTTATTTTGAAAATGAACGATTTGATTTCAGAAAGCAAATTCGAAGAAAGAT
CCCACTGGATATGTGAACAAAAAAGAAACAAAATGGATCAGATGCAAGCAGATCAATATGCATCTACCTCTTGCGCAATG
AAACAAAAAACTGATCCAGTTAAACAATGTAATATTGGATCAACAGCGGTATATAGAATGACAATTGAAAATAAATGGAA
TTTATTAAATCATTGGAAAGATTGGCCGGGCAAAGACGGGTATTCTCCTAAAATAACTCAATATCTCGGAGCTTCTCATT
CAAATGAGTTTAATATTTATCATCTTGGTGGAATGGCCAATGAAGGAGTTTCTCAGGTATGTGGAAACGGGAGCTACACT
GAATTGCAAAAGCAATTCTACAAAGCTGGGAACGGGATCATGACAGTTATATTGGCCAGGTCACTAGATGCGTCATCAAA
AACCAATAAAAGATCAGCACTGATTGTTGTAAATGCAACTCAACATTTGGTTAGTTTTCTTGCAAGACTTCATCCAAGTC
CCGATTGGTGCACAGGTCTAAGTAGATTTGATCTTTGTAACGGCTCATGCAAATGGCAAGAAAAATTTATCGTCCATTTA
TATCCTTGGGATGCCGGTGTTTACAGTGGAGAGACTTATTTAAATAAAAACGAGAAAACTAGACAGACTCCAATATGTCC
AATCACTAGCAGTTGGCCAAAGGATAACCCTTTCACGGTGATGAACGGAAAAATCAAAAATATCGGAGTGGTAACCTTCC
AATTAATACAAGAATATAACAAAAATGATAACCAGTATTGTAATTCTATTAATTCAATCCTTCAAGACGAAGCATACCAA
AATGAACCGAAATCCAGTGCTCTGAAATCAAATCCAGATAAAACCAGGCATTGCTTATTGTCAGAATGGAGTGAATGGAG
CTCTTGTTCAGAATCTTGTGGTAAAGGTGAAAAAACTCGTTACCGAAAACTGCTGAAAGGAAAATCCGAACAATGCATTG
AATCTTCCCTAACTAATACAGAGAGCTGTAGCAGTAAATGTTCGACAGTAATTTCATTCGAAACATGTTCGTTTCGCTTG
TGGGGCCAATGGAGCCCATGCAATGCTACTTGTTCCCAGAAGGGAATTATTCATAGGGAACGTGTTTTTATCAATGAACA
GGAAAAACACGAATGTATGCAATATTCCGAAATGAAAGCCTCTCTTGAATGTGCATTAGCAAGCGAGAGGTGCAAACCCG
ACGTGATCTGCAAGGAATATGTTTATCCTGGAGATTCGTGTTCTTGGTCTAGCCCATCACAAAGATTTTATTACGATTTT
TCTAGTGGAACTTGTAATGCATTTATTTACAAAGGCTGCTATGGGGGTCTGAATAATTTTCACACCTACGAGGAATGTAT
TTTACTCTGTCGACCAAAATTAAAAGAAACAGATCAAAGACGATTTAATACGCCAGAAACTTATCCCAACCGATGCGGAG
TTTCGATGACATGGGGAATCTTTTGTGATAGTCAATCTTCATCAAACCGTTGGTACTATGACAATATTAATAAAGAATGC
TACCGATTTCGTTTCGGGGGCTGTCGAGGGAATGCCAACAATTTCCGAACTAAAACTGAATGTCAAAATGTTTGCTTGAG
AGAAAAGTCTACTTCAACAACAACGACAACGATCAGTCAGCCAGTAACGGTAAGAAAAGTCTGCATCTATTCCAACTGGT
CGGACTGGAGTTCTTGTGATAACAACTGTCAAAATCAACAAACAAGAATAAGATTTTTAATTGAAAAAAATGACGACTGC
ACCGAAAGTATCAGTGAAACAAGAAAATGTCAATCAGAGAAATGCAATTCGCCAAATAACGGGTTTATTTCACTAAGGAA
TTGCAAGTATTCGAGTTGGTCGGAATGGTCGGCTTGTTCATCAAGATGCGGTCCTGGCGTTAGAGAGAGAAAGCGGAAGA
AACTTTCTCGTAATTGTAAACCGGAATTTCCGCAATTTACTGACCGAGAATCATGTTACAATGCTTGTGCTTATGACTGA
ATCCGGTAAAGAAAAAAAAACTGTCAATGTATTTGTGTCATGACATTTTACCTATTGTGCATTTCATTTTGTGAACTTTG
TTTGGTTTTTGATTTATTTTTTCTTGGTGTTTTTCTCTAGAATATGATTTGTT
>SMED30019695 ID=SMED30019695|Name=Cathepsin L|organism=Schmidtea mediterranea sexual|type=transcript|length=1216bp
TTGAAATTCTGCATTGATGGGAATTGCGTCGTAATAATCACGCGCCGAATATCAAATGTATAAATAAGATATTTGTTCTT
CACTTAATATAATTATAATGTTTGCGATGGGTCCTATCTTGATATGTTTATTATTAGCAGTGATTTCAGAAGCCAGAACC
ACTTATAGCCATCACAAATCAGTTCCAAGTGATGACATTTCTAGTAAATTTGAACTAGAAAATATGGTATCTCTCTTGTC
CCGTTACGACGGAGAATTCCAAACCTTCAAACAGCGATATAATAAATCTTATAGTTCCATGCATGAAGAGAATTCCAGAT
TTTTAAATTTTATATCATCTCTACTTGAGATCAATCATCACAATCAACTTTATGAAAACGGTCAGAAATCATACGAGATT
GGAATCAACCATTTGAGTGATTGGAGTGATGAAGAACTCGCTCAACTGACTAAATTGAAACAATCAAACCTGACGGGCTC
ATCATATTTATCATCCAGTGTGAAAATTGACCTTCCTGATTCATTCGACTGGAGAGAGAAGGGTGCAGTTACTGAAGTTA
AAGATCAAGGAAAGTGTGGATCATGCTATGCATTTGCTGCTACTGGCACTCTAGAAGCCCAAATCTGGAGAAAGACTGGG
AAATTGATCAGTTTATCAGAACAACAAATTGTTGATTGCGATACTAATGATAACAATTGTTACGGTGGATTTGTACAAGA
CGCGTTATCATACATCAAGTCAAATGGAGGCATTGAAAATGAGGAAAATTACCCTTATGTTTCCGGCAAAACAGCAGATA
TGAACCCAAAATGTAATTTCGTAAAAAGCAAAGTAGTGGCGACTGATACTGGATTTGTTAAAATGCCAGCAAATGATGAA
GAAAAATTAAAAGAAGCTCTTGTTACAATTGGCCCTATTTCTGCATTATTATCTGCTCCTAAATCGCTTATAAAATATTC
AAAAGGAATTTTCTACGACAAAGAATGTGAAAAACTAAGAGTTAATCACGCTGTTCTGATAGTCGGATATGGAACAGATC
AAACTTCCGGGACACCATATTGGATAATTAAGAATAGTTGGAACGCATCTTGGGGTGAAAAAGGATATTTCCGGTTACCC
AGAAATATAAATGCTTGCAAAATAGCGTCAGTGCCAGTTTATCCTTTGGTTTGAATTAAAATGTTTATGTAATAAATATA
TTTTTTTGTCTCAAAA
>SMED30029077 ID=SMED30029077|Name=Macrophage-expressed gene 1 protein|organism=Schmidtea mediterranea sexual|type=transcript|length=2367bp
TAAGTAAAACAAAAATTCGAAATTTGTATATTAATGCGAACAAAGTTTAAAAATTTATTGTAAATGTATATTCAAATATT
ACTAATAATTAGTCTTTTCAAAATATTAGTGAGAGGATCAAATGAAGTCAATACTATTAAGGATGCGTATCAATTGTGTC
AGTTGATGCCCAATCAAAGAACGTTCGATGTTTTACCTGGATCAGGATGGGATAACCTGGCCAATATCGGCATGGGAAGG
ATACTTGATTTAAACTTTTCGAAGTGTTCATTTTCCAATGACAACAAATATCTGATCCCGAATCAACTGGCCATTTATCC
AGAAAAATCTTCCCAAGTTGAAATCTCAAGTGAAATATTCCGTCATTTCTCAAATTACACATCTATGTCTTCGCAATCTA
TCAATGTTGGTGCTGAAGGTGGCTACGGAATATTTAAAATTGGGGGATCTTTTTCCACGGAAAATACAAAGATCAAAAGT
GATCAGATGGAAACGAAATCCATTTTCACCAGAGTTCAGTTGCGATATTTGGGGTTCCAGTCAATGTTTGAGCCGGATAC
GCCTCTCTATCCAACATTCAAGAACAGAATTATGGACATTGCTGGATTAGTTGTTGAAAATAAAGACTCACTTTATCCTG
AGTATTTGGCTGACATGATTGTTCGAGACTACGGGACCCATGTAATCCGTGCTATAGATGCTGGGGCGATTTTGTCAAAA
ACCGATGCTTTGAATTCACAATACGTAAAGAAATATGAATCGGATAGTTCCACAATCTCCGTTTCTGCGTCAGCATCTTT
TCTTAGTTTGTTTAAAATAGATGGTAAATACCAAACTAAAACTACACAAACGAATATTGATGAGTTCAATCACAACGTTG
TTAAAACTCACATTGATACGGTCGGAGGTGCCATATTTGGAGCCAATACGTCTTTGTCCGACTGGGAAAATTCTGTCATG
AATAATTTAGTTCCCATAGATCGGAGGGGTGAACCATTATACTATGCCATTACTCCTCGTGCCTTGCCAGAACTGGATCA
AGAAACCGTGCACAAAATTTATGTCTTAATAAAGAAAGCAATTCTACGATATTATACAGCCAATGGCCGAATTGGATGCC
TTGATATATATTCTCCAAACTTCAATCCGGATGCCAATATTGATTCTGGGAATTGTGAACATCCGTCTGTGAATTTTACT
TTTGGAGGTACTTATCAAGTTTGCGGTGGTCCTGCATGTGGTCCATATGCTCAAGTAAATCCATTAACAGGAGGGTTTTC
TTGTCCCTCTGGATTTATTTCGATAAAACTTCACAGTGGCTTAGTTCGATGTGACGAATCATGTCATGGGTGGTGGTTTT
GGAGAAGCTGCGATACAACTTGCTCTTATTTTACCACTTTTTGGTGTTATCCGGTGAATATCGGCCCAAATTACAATGGA
TATCAATTTGGAGGAATTCGTCAAGGAAGTTGTCCCATTGGATATATTCCACTCAAGGTCGGATTAAGCATAGAAATTTG
TGTGACAGTTGATGCTGACCCGTTCAATCCATATGCTATTAAATTTGGCGGCTTTTTCAGCTGCAGTGCCGGAAATCCGC
TAGCTCAAGAATATGTTAAAAACAAACCAAAGTTATATTCCAGTTCGAAAATGGCAGATTTAAAATATTGGCCTAAAACC
TGTGCCAAAGGTTATGTGGCTCACATTTCATCAATTGAACAAGGCTGTCAGATTTCATACTGTACAGAGACAAATAAAGT
TTCTTTAGGCCCTTTACCTCGGATAATTAAACCTCCATTTATTTCAATTGATTACAATCCAAACAGCACGAAAGTTTTAA
TATTTACAACCAAAAACGGTGAAACGTTTATAAAAGATAACGTTGGATCATGGAAATTAGAGGACATTCAAAGAGAGAAA
AGAAATATGTTAGTTGGATCAATCGTTGGAAGTTTTCTAATGGCTGCCTGCATCTGTTTGACGATTTTCTTCATTCAAAG
AAGACATTATAAACGAATGATTGAAAAGTTAGATTATAAACCATTAACAAAGAATGAACAATTGGCGGACTACGGCTCAA
TAAATAATACAGACAGAGTGTAATTAAAACAACAAAATTAAATGTTTTTCAAACATTTTTAGCTTAGCTCATGAAAACAA
TTTACATTGTTCACAAAATTTATTTGCATTTTTTATCAATTATTGTAATTGCTTAATGTCGTTAATAATTACATTTTTAT
ATGGAAATATATCTGATTTCATTATTTTTTCAAAATGATAAGATGACTATTTAATTTTGTTTGATAGCAAGAGAACAAAA
AATTTTGTGATTAATTCTGATGATTTATTGATCTATTTTCATTTGAC
>SMED30012382 ID=SMED30012382|Name=Macrophage-expressed gene 1 protein|organism=Schmidtea mediterranea sexual|type=transcript|length=2261bp
CGCAGAGGTCCCTCTGGCAACACCGAAGGAAGTGTGTGACAATGAAACAAATCAAAATTAAGATTTTACCATTTTGAAGT
TGAGTCAAATATTTCATTGATGACCAATGCAGTGCAAATTAAAAGTTGACGTATACAAAACATACCACAGTAAGAAGGCA
ATTTCTTTCCGAGCATCCCGCAAATAATCCCAAAAGAGATATTGCAATTACTATGATCCCAATAGCCAAAACAAAAGTAA
CTGATGGAAAAATAGAATATCTTTGTGCCAAATTCATGCTGAATCAGTAGTTGAACCGGAAGATATTATAAAGCAAATGA
ATCTGCCATCCTCTAGAATTGTGCATAAATGTCACTTGACCAAAACTCAGTATCCTTTTAATGTTTTACCAGGATCAGGA
TGGGACAATTTAGCAAACAAGGAATCGGGACAAGTTTACAAAATTTCATATGATAATTGCTCGGGATCAAACGATGGACG
GTTTTTAATCCCCGATTCAATCATTATAACTCCATACATGAAATCCAAAGTCCAAATGGATTCAGATATGGTTTCTCATT
TCTCAAAATATACATCAACGACTTCTAAATCTATTAATCTTGAAGTCAGTGCTTCTACGCCAAATTATGAAATAAGTGGT
TCATTTTCATGGGAAAAGAAAGATATTAAAGAAAATCAAATGAAAGAAAAATCATCTATATCCAAGGTTCAACTACGATA
TGAATTGTATCACGTCGCATTTCAACCTGATTCAAAGTTGGACCCTATTTTCAAAAACAGAATTATACAATTGGGAGTTC
TGATGGAAACGCAAAAGAATTATCTATATTCCCAGTATTTAGCCGATTTGATAGTTCGAGATTATGGAACCCATGTTATC
AGGTCTGTGGATGCGGGTGCAATTCTGTCAAAAGTTGACATACTGAAAGCAGAGTATTTGTCAAAACATAAGGGCTCCCA
AACGACAATTTCATTAGCAGCATCTGGAAGCCTGAATTCAGCAATTGGCTTAAAAATTGGCGGCTCAAATAAAAATAACG
AATCAGAAGTTCGCGAGTACAATAAGAATTGTGCTAGCTCAAAAATTAATACAATTGGCGGTGGGCCATACGGATCCAAT
TTTCAATTAGCTGAATGGGAGAAAACTATTCAAGAAAATTTGGTTCCAATTGATAGAATCGGAGATCCGCTATTCCACGT
TATAACTAAGAGAAGTTTGCCAGAGTTTGACGATATAACGGTTTATAAAATTGGAAATTTAATTAAAGACGCAATTACAA
GATACTATAAAATAAATACGCGATCGGGTTGCATGGACATAAAATCCGATAATTTTGACGTAAATGCAAATGTTAATAAT
GGGATCTGCCAAGACTCAAAAATCTCATACGTTTTCGGAGGTGTCTATCAAACATGTGAAGGAAATTCGTGTGAAAATTT
GATTCAAACCAACCCAGCCACTGGTTCAACTTCTTGTCCAGCGGGATATAATGCAACCTTAGTCTATCACGGCTTATATT
CTGAGCAAACGATTTGTACTTATTATGTTTTCTCTACTGTTTGCAACAATTTCCAAACATATATATCTTCATATTGGTGT
GCTCCTGATCCGAAAAAAAATCCGGAAAGTGTATATTTATTCGGTGGGTTTTACTCAAAGGTTTTCCCTAATCCAATTAC
AAATTCTTTCACTTGTCCACAAAGTTTCATTGCTCTACGTTTCGGTCATAGTTCATGGGTTTGTGTTACGTCAGATTCAG
TGCTTGGGAGAAAATTCAGTTTACCTTTTGGTGGATTTTACAGCTGCATGAGTGGTAACGCGCTGGTGGCTGATGCATCA
GCACGATCACAGACCTGGGCAAAATCTTGTCCCCCAGGCTATTCGGCCCACTTCGCATTTTTAGAAAACAGTTGTCAAAT
ATCGTATTGTCTAAAAGCCAACTCTCTTAAAGGTATCAAAGAAATACCTCTTTACCGGCCTCCATTTATTACGATGGACA
ATGGGCCTAATGTGACTGACGTAAATAAATTCAAAACTTCATCTGGAGTCAATCTTGTTCTCTCCAAAGATGGACTATGG
AAGAGGAAACAAAAGAAAGTTTTCATGAAAACCCTAATAGCCGTTATCATTCTTGGTATTTTGATAGCCGTTTGTATAAT
AATGAATGGGGCTTTCGGTTTGAGGTTATTGATATCAAAAAAACAAAAGTAGAATAAAAGTATATTCTAAAATTCATTTT
GATGAATAAATGTTTTGGGTT
>SMED30017598 ID=SMED30017598|Name=Cathepsin L|organism=Schmidtea mediterranea sexual|type=transcript|length=824bp
TTCAAATATGATTATCTCATATTATATTATATTGCTTTATTTTTTCATCCTTCTGACTGAAGCATCACCCGTACTTCGTC
TTTATTCTATTGTTTCTAATCTAACATTCTCGCCTTTCATTAGTCAGTTTTTAATAACTGATCTTCCTGAATCGGTAGAC
TGGAGGCTGAAAGGAGCAGTTACTGAAGTTAAAGATCAGCAAGAATGCGGCTGTTGTTATGCATTTTCAGCTATTGGAGT
AATAGAATCTCAAATTTGGATAAAAACAGGAAAACTTGTAGATTTATCAGTTCAACAAATTTTAGATTGTGATGGAAAAT
CTCAACAGTGTCATGGTGGATTTCCCGACGACAATTTCAATTATATAACTAAAGCTGGAGGAATCGAGACTGAAAAGACC
TATCCTTATGTTTCTGGTTATAATAGTCGGGAACTTTATAAATGTAAGTTCAATGAAACGAATTCCGTGGCAGCTGTAAA
AGGATTCGTTCAAATTCCAAAAGGGGACGAAGATGCATTAATGGAAGCAGTGGCAACAGTAGGACCAGTCTCTGTCAGCT
TGTGTGTTTCCTATGACCTTGGTTCTTACACTTCTGGGATTTTCAAAGATAAATTGCCTGATCATAATGAGTTTAACCAC
GCTGCTATCGTTGTTGGGTATGGAAATGATAACGTAACTGGGACACCATATTGGTTGTTAAAAAATGCTTATGGTAAAGA
GTGGGGAGAAAATGGATATTTTCGCATTGAAAGAAATAAAAATTCCTGCTACATTGCTCAATACGCTTTGTATCCCATTG
TTTCAGTTACGTAAGAATAAACTC
>SMED30013909 ID=SMED30013909|Name=tud-1|organism=Schmidtea mediterranea sexual|type=transcript|length=2414bp
ATTTAGAAAATGGAGGCAACTTGCGATGTTTTTTTAACGAATTTACCTAAGTCGATTAATGAAGTAGTTGTGAAGAACTT
ATTTTCTTCTGCTGGGAAAGTGTTAAAAGTGAAAATAATAGATGACAAAAGAAACGAAAATCAGAAGCTATGCTTCGTTG
GATTTCGAACTATGGATGAAGCATTAGAAGCCGTCAGAATTGCAGATAAATCTGATGCGTTTGGTGATCCAATTAGAGCC
AGTATATCAAAGAAGTGTTTAGACGGAATGGAGAATAATCAAAATAGATCAAATAATAACAGAAATGATTCAAGTTATAA
AGGAACTGATTCATTTTCAAGAAAGAGTGATTCTAATTTCAAGGATAGTGAAAAGTATCCAACAAACAATGGTGACTTTG
CACAAGTAAATCGTTTAAACAACGTTGGAAGAGGAAGAGGAATCCAGACTAAAATTATTAGTGGTCCACCTTCATTCAAA
ACTAATGAAATTAAGTTAAACGATATTAAACTTAAAACAAAAGAACTGATTGACATCGAAATCGTCGATTTAAGTAGAGT
TGACATTATACTTATAAGGCCAGTAAAGTCAACGGAAAATTCAAAAATTGAAATAGCAGATTTATTGTCACAAATAAATT
CAAATAAATCGAATCTAATGCCAGTTGACTGTCCTAAAATAGGAGATTTTTGTCTTGCAAAATGGACTGAAGATGATTGC
TATTATCGGGCTGAATTGTTAGATATATTAAAGGATGGATCTTTCAAAGTTACTTTTGTTGATGAAGGAACTTCGGGTGA
TATTGTGACTGATTTGAAACACATTCCGACTGGTGCTTTACAGCAGTATCCTAAACAATTTTTGAAGGTTTCAATCGGTT
GCTCAGACGCAAAAGAATTAATGTCATTTATTTTACAAGAGTCTGAAGGAAAGGTGATTAAAATTTATTGGCTCGGAGAA
TATTGTGATTCCGTGCCTGTTGTAAATTTATTTCTGAATGAAGATGAAATTGTGGAAAAATATACCCAAATAAATAAAGT
AAATATCAGGAAAAAATTCTCACAGACATTTGATGATCTCCCAGTACATAACCTCGAAACTAATTCACATTTTTCAATAG
CACCATCAATGTATAAAAACCGAAATGATTTTATTCTTCAGATGAAAACTTTCTCGGTATCTTTAAAGCAAAAAAAATGT
CCTACGGTTTTACCAATTCCTGGTGATATATGTATATATGAGAAAAGGAATGCTAATGGCGAATCATTTCTTCGTTGTGT
TATATTAAGGCGCCAACTTTTTTCTTTTGACATCGAGTTAATAGATATCGGTGAAAAAATAATGAATGTTAATATTGATC
ACCTGTACACAGTCGAAGATATTCATGTAAAAGACGCTCCCTGTGCTATTACTTGCGATTTAGATAGTTCTATTAATATA
GATGGTATTAAGCTCCCTCAAGATTTGATCGATGGTGTTCTCTTTAATCTTAAAAAGTACAACAAAAATAATAAATACAC
TGTAAATTTCATTTTACCACAAAAGAAAGCTTATGAATATGCAAAAGTATCATATAAAGATTTGATTGGTAAAAGCTTTA
AAGTAATAATGTCGCACGTTGTCTCTCCCATTAATTTCTTTATTCAACTTGATGATGGAGAGCATCAGCTTGCACTTGGA
GATATTCAAGAAAAATTAAATATTAATTATAATGGATCACCATTCAAAAGTGCTAGCGATGTTAAAGAAACTCAGGCAGT
TATTGCTCCTTTTATTGTTTCCGGAAGTGATGCGGAACTTTTTCGGGCTTATGTTTTAGAGAAACGAAAGGAATCTCCAC
AAATTCATGTATTGTTTATTGATTATGGTAATGATGAATGGGTGAATTATGAAACTTTACTGAATACAAATGAAGAATTA
GTTTCTAAACCAGGTTTCGCAATACATTGTAAATTATCTCTAAATATTAAGAAGGAAGCGTGGAGTCACGCTATTGAATT
ATTTAATGATTTAGCTGGTTATACTGAAAATGACCAGGGGACGTTTGTAATGCAGGTTATTAGTGTCGATGGTGATTCGT
TAGTTGTGGACTTGATTTCAGAACTGAAAGGTTCGTTGACGGCCATCTTAGAAAAAAGTGGTACATGTGTAAAGTCCAAT
GAACCAATATGTAATGGAGGAGGACCCCAAGTTAAATTAAATGGCGTTTCAAATGATAAATCTCAATCAGCTATTCTTGA
AAATTTATCTGAAGGAGAATTATATAAAAAATTTATTGAACTAGAACGGAGATTAAGTGCTTTAGAGATATTGATGAAAT
AATGCTACCCGCGCAATTAGTATTATCAAATTATGATAATCTAGTCTTGATGTGTTTAAAATTTTGTATTTTAACATAAA
TTTATTCCGTGTTG
>SMED30001027 ID=SMED30001027|Name=Cathepsin L|organism=Schmidtea mediterranea sexual|type=transcript|length=1243bp
TAATAATTTGCACATCAAACCGTAATAATAACGGATCCGTACAATTTCATCAATATAAAATATTACACAACAAATGCGAC
TTGCATCGACAATTCATATAAAAATCTCCAACTTTCACTACTATTCATAAATTACAAATGATCTCCTCAGTTGTATTGTT
ATTTTCTATCTCAGTCGCATTCCTAGCAAATGCCGAAATCTGGGAACCAGATCGAAACCTTTCAAACGTCCATCAGAGTC
ACGCTAATACAAAGTTTTCCAGGAAAATTTCATCTTTGATGGTGGATTTCAAGTCAGAATTCGCCAAATTTAAATTACAA
TATGGAAAGGTTTACGAGTCAATTGAGAAAGAAAACGAAAGATTCAACAATTTCATATTATCTCAAGTTGAAATAAACAA
TCACAATCAAATGTTTGAAAAGGGTCACAAGAAATTCCAAATGGAAATTAATCACATGAGTGATTGGACTCCAGAAGAAT
TCAATCAACTTAATAGCCTAAAGATATCTAATCTCACCGGTTCGACATATTTGCCATTGAACATAAAGCCTGGCCTTCCA
GAGTCTTTTGATTGGAGAGACAAAGGTGCAGTTACTGAAGTCAAAAATCAGAAATATTGCGGTTCGTGCTACGCATTCTC
TACGACAGGCGCGTTAGAAGGTCAACTATTCAGAAAAACCGGAAATTTGATAAGTTTATCTGAACAACAAATTGTTGATT
GTGATCATGTTGATTATGGTTGTAGTGGTGGCTTAGTACCAGACGCTTTCAATTACATCAAATACTCCGGAGGAATTGAA
TCCGAACAAGAGTATCCATATGTATCTGGATTGACAAAGAAACCAAATCCAAATTGTAATTTCAAAGAAAAAAAAGCAGT
TGGAACAGATACCGGATTTGTGAAATTGCCTCCTGGAGATGAAGAGGCTTTGAGAGAAGCTTTAGTGACAATAGGCCCGA
TTTCGATAGCGTTTTCCGCCACTGAGCGATTCAAACATTACAAAAATGGAATTTACCATGATGATTGGTGTTCAAATGTC
CGTATCAATCATGCAGTTTTACTCGTCGGATATGGAATGGATGAAGAATCAGGAACGCCTTATTGGATAATAAAAAATAG
TTGGGGACCCAAATGGGGAGAAAACGGATATTTCCGTTTCGAAAGAAATAAAAACATGTGCAACATGGCATACAATTCAT
ACTATCCATTGGTTTGATTGAACGACTTCTTATGAGTGTGTGC